2009 Summer Project Week-FastMarching for brain tumor segmentation
Key Investigators
- BWH: Andriy Fedorov, Ron Kikinis
- GeorgiaTech: Yi Gao
Objective
We will investigate the currently available algorithms and tools for fast marching segmentation, and study their applicability to the problem of segmenting meningiomas in contrast-enhanced brain MRI.
The key requirements to the segmentation approach are naturally accuracy, but also simplicity of initialization and acceptable speed for interactive analysis.
Approach, plan
We will evaluate the fast marching segmentation methods developed by the GeorgiaTech collaborators on clinical meningioma MRI data. A set of anonymized images representing meningiomas of various appearance will be used for testing. Based on the results of the evaluation, decisions will be made about the most appropriate method to be implemented as a Slicer module.
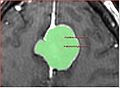
The major issue with the fast marching segmentation available in Slicer3 currently is underestimation of the structure. It leaks before conforming to the boundary of the structure. We would like to investigate if this issue can be remedied.
Progress
- FastMarchingSegmentation module by Eric Pichon was ported into Slicer3 (available in Slicer nightly build)
- 4 representative meningioma datasets are in Sandbox http://svn.na-mic.org/NAMICSandBox/trunk/MeningiomaSegmentation/, we would be interested in evaluating and comparing any available segmentation methods for this application
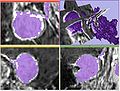
- evaluated itk-SNAP and VMTK-based level set segmentation from Daniel Haehn on the test data. Main issues are accuracy and parameter selection (itk-SNAP).
- ITK-based segmentation with geodesic level sets complete, but en plaque tumors cannot be segmented with that technique based on initial experience.
- ITK level sets time acceptable, but need to work to make tools more interactive and intuitive
Meningioma segmentation challenge
You can evaluate your segmentation method on the meningioma tumor segmentation, and we would appreciate if you contribute the results and details of your segmentation. Sandbox directory with the input data has been setup to keep the results. Instructions are in the README in MeningiomaSegmentation Sandbox directory:
The images in the Data directory contain volumes of interest with meningiomas, and the binary segmentation of tumor in each image. The volumes were supersampled 2:1 in each dimension from the original data spacing. Linear interpolation was used for grayscale data, and nearest neighbor interpolation -- for segmentations. If you want to contribute a segmentation result, please create a new uniquely-named directory, and save the segmentation results together with a README file, which would hopefully include the following sections: ** Method: how did you perform segmentation? ** Parameters: how can your result be reproduced? ** Results: where are the segmentation results saved? ** Execution time: how long did it take to run your method? ** Parameter selection: How difficult was to find the parameters that you used to achieve the segmentation result? Did you use the default parameters? ** Platform: what are the characteristics of your machine, is your code optimized? (rough estimate is ok) ** Contact See the SegmentationMethod1 directory for an example of such description and segmentation results. Please contact Andriy Fedorov (fedorov AV bwh DOT harvard DOT edu) with questions or comments.