Mbirn: Defacer for structural MRI
- Short Description
This package contains an algorithm for removing identifiable facial features (eyes, nose, and mouth). This algorithm, which is still under the validation phase, locates the subject's facial features and removes them without disturbing brain tissue. The algorithm was devised to work on T1-weighted structural MRI; it produces a defaced structural image, and an image of the applied mask. The code mri_deface produces a facial mask. The code mri_mask is then used to apply the mask to the original volume, resulting in a defaced structural image. For images in other modalities, the mask can be co-registered to the image before application.
Keywords: HIPAA, data sharing, de-identification
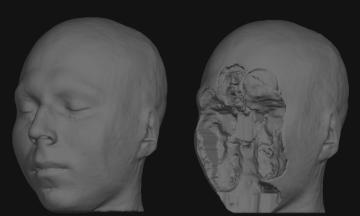
De-identification results. Surface rendered reconstructions from a structural MRI scan using the raw acquired data (left) and the de-faced results (right).
- Longer Overview
- Due to the increasing need for subject privacy, the ability to deidentify structural images is crucial. A program was developed that uses models of non-brain structures for removing potentially identifying facial features. The facial features of ten subjects were manually labeled, and an optimal linear transform was used to build an atlas of face membership. When a novel image is presented, the optimal linear transform is computed for the input volume (Fischl et al 2002, Fischl et al 2004). A brain mask is constructed by forming the union of all voxels with nonzero probability of being brain. This mask is then morphologically dilated to yield a binary volume, the nonzero values of which indicate the presence of brain within x mm. Finally, all voxels that are outside the mask and have a non-zero probability of being a facial feature are set to 0.
- The algorithm was applied to 342 datasets that included two different T1-weighted pulse sequences and four different patient diagnoses (depressed, Alzheimer’s, and elderly and young control groups). Visual inspection of the defaced images showed none had brain tissue removed. In order to quantify the effects of defacing on a volume, 16 of the above datasets were bias corrected with N3 (Sled et al 1998), defaced, and then skull-stripped using either a hybrid watershed algorithm (Ségonne et al 2004, in FreeSurfer) or Brain Surface Extractor (Sandor & Leahy 1997, Shattuck et al 2001); previous study of skull-stripping algorithms suggested that these techniques may be the most adept at automated skull stripping for the pulse sequences and patient populations employed herein (Fennema-Notestine et al in press). Our results suggested that the automatic defacing algorithm is robust and efficiently removes non-brain tissue that would have similarly been removed via skull stripping. Additionally, defacing did not unduly influence the outcome of the processing methods utilized; in some cases skull-stripping was improved. Analyses support this algorithm as a viable method to allow data sharing within large-scale multi-site projects. A manuscript detailing these results is nearing completion for journal submission.
- An additional consideration is whether the defacing algorithm renders the individual unidentifiable. Although MR images lack relevant identifying features (such as hair, eyebrows, and skin pigmentation) or may be of poor quality, it is still possible to identify a subject, particularly if the subject is familiar (Bruce et al 1999, Burton et al 1999). This is particularly troublesome if the subject is participating not as a normal control, but as a representative diagnosed with a particular illness (e.g., HIV, depression). The purpose of a defacing algorithm is to protect the identity of the participant. Face recognition studies have shown that these internal features (eyes, nose, mouth) are more important than external features (hair, ears, face outline) for recognizing familiar subjects, whereas both feature classes are of equal importance for identifying unfamiliar subjects (Clutterbuck & Johnston 2002). Therefore, we are conducting a behavioral study to determine whether defaced images may be identifiable to both individuals familiar and unfamiliar with the person depicted in the 3D rendering of the MR image.
- User Manual
- A README text file with use and installation instructions is provided in the download
- Registration and Installation Information
- Download from: Download (linux/mac)
- Obtain Freesurfer license key from: http://surfer.nmr.mgh.harvard.edu/registration.html
- Set environment variable FREESURFER_HOME and copy the license key there. The programs mri_deface and mri_mask work if the freesurfer license key is found in: $FREESURFER_HOME/.license
- Note: you don't need to download or install Freesurfer to use mri_deface or mri_mask, you just need the Freesurfer license.
- References
- Bischoff-Grethe, A., Fischl, B., Ozyurt, I.B., Morris, S., Brown, G. G., Fennema-Notestine, C., Clark, C. P., Bondi, M. W., Jernigan, T. L., and the Human Morphometry BIRN (www.nbirn.net), June 2004, A technique for the deidentification of structural brain MR images, Human Brain Mapping, Budapest, Hungary.
- Bruce V, Henderson Z, Greenwood K, Hancock PJB, Burton AM, Miller P. 1999. Verification of face identities from images captured on video. Journal of Experimental Psychology: Applied 5: 339-60
- Burton AM, Wilson S, Cowan M, Bruce V. 1999. Face recognition in poor-quality video: Evidence from security surveillance. Psychological Science 10: 243-8
- Clutterbuck R, Johnston RA. 2002. Exploring levels of face familiarity by using an indirect face-matching measure. Perception 31: 985-94
- Fennema-Notestine C, Ozyurt IB, Brown GG, Clark CP, Morris S, et al. in press. Quantitative evaluation of automated-skull-stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Human Brain Mapping
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, et al. 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33: 341-55
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Segonne F, et al. 2004. Sequence-independent segmentation of magnetic resonance images. NeuroImage 23: S69-S84
- Sandor S, Leahy R. 1997. Surface-based labeling of cortical anatomy using a deformable database. IEEE Transactions on Medical Imaging 16: 41-54
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, et al. 2004. A hybrid approach to the skull stripping problem in MRI. NeuroImage 22: 1060-75
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. 2001. Magnetic resonance image tissue classification using a partial volume model. NeuroImage 13: 856-76
- Sled J, Zijdenbos A, Evans A. 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging 17: 87-97
- Technical Contact
Support is very limited but we will do our best to try to answer your questions. Please email your constructive comments to: Bruce Fischl (fischl@nmr.mgh.harvard.edu)
- Acknowledgements
- MRI Deface is brought to you by the Morphometry Biomedical Informatics Research Network (BIRN, www.nbirn.net), a project funded by the National Center for Research Resources (U24 RR021382), at the National Institute of Health, U.S.A.
- Special thanks to Amanda Bischoff-Grethe for testing and validation work.