October 08 2009 Meeting: VT-NAMIC Revised atlas construction workflow
Contents
TODOs
Software
Workflow
Template Tommy (deprecated)
Step 1: Prepare the data using the instructions -- LINK
Step 2: Gradient anisotropic diffusion. Tools: Slicer GAD module. Parameters: time step: 0.0625, iterations: 5, conductance: 1. Note: Should try with and without GAD. Sandy suggested GAD can cause troubles for EM.
Step 3: Initial registration 6+9+12 DOF registration. Tools: BRAINSFit svn r301. Parameters: default except CenterOfHead initializer with ROIAUTO sampling region. Comments: failure to recover the pose for Hugo and Valentino (both have severe head tilt in all three planes). Execution time: ~2 min per subject.
Step 4: Manual pose alignment for Hugo and Valentino. Transforms "hardened" in Slicer MRML, volumes saved and re-registered same way as described in Step 3. Registration issue resolved. Tools: Slicer Transforms module, BRAINSFit.
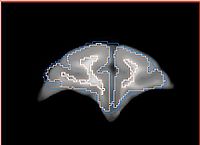
Step 5 -- Ginger: Manual ICC for Tommy. Tools: itkSNAP, manual editing by Ginger. Tried Xiaodong's SkullStrippingModule, no luck.
Step 6: Editing of ICC label map. Tools: Slicer smoothing label map module. Parameters: 10 iterations.
Step 7: Manual mask dilation, so that each brain of each subject following registration in Step is contained. Tools: Slicer Editor module. Parameters: 8-neighbor, repeated 3 times.
Step 8: Resampling of the ICC to each subject. Tools: Slicer ResampleVolume2 command line module. Parameters: NN interpolation.
Step 9: Bias field correction with ICC. Tools: Slicer N3MRILightCLI extension. Parameters: default values except number of iterations increased from 50 to 100, convergence threshold decreased from 0.001 to 0.0001, ICC label from Step 8. Execution time: < 2 min per subject. Comments: There are some parameters in N3 which we understand and can play with. Subjective (?) trying to evaluate if the result is better. Note: should also try not correcting for bias field outside EM.
Step 10 -- we are here right now: Pipelined registration (6+9+12+BSpline). Tried: (1) ICC ROI from step for moving floating, BSpline step segfaults; (2) ROIAUTO (same as in Step 3), BSpline completes, but resampled volume does not make any sense. AF: I suspect BSpline registration may not be able to handle non-axis-aligned volumes. Comments: When resampling, do not forget to use higher order interpolator. Debugging data: File:Tommy-Calvin-bspline registration.tgz
Step 11: Average all subjects to get average template.
Step 12: Manually edit ICC labels for each subject, skull strip here or after Step 9, edit and repeat all steps to improve registration.
Remaining steps:
- Segment GM/WM/CSF+Putamen/Caudate/Hippocampus in the averaged template.
- Invert the deformation field from Step 10 (Step 12?), and map the segmentations to the individual subjects.
- Manually edit each subject labels. Map back to the average template and calculate the probabilities atlas.
- Run EM Segmenter to get the bias field. Repeat again?
Template Gucci
Following the discussion during Oct'09 meeting, the agreement is that Gucci is the best subject to be used as a template for atlas building (see meeting notes). The current progress in atlas construction will be reported here.
Transforms, scripts and other relevant material (except data) will be maintained on the NAMIC Sandbox here: http://viewvc.slicer.org/viewcvs.cgi/trunk/VervetAtlas/?root=NAMICSandBox
Step 1 Prepare the data using instructions. Result: <Subject>-raw.nrrd
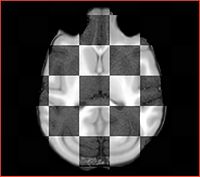
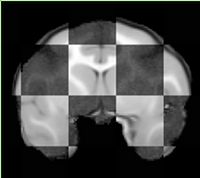
Step 2 Discard the directions matrix from the input data using DiscardDirectionCosines Slicer extension. Images with non-orthogonal orientation result in poor performance of B-spline registration at a later step. Result: <Subject>-axial.nrrd
Step 2 Segment Gucci (template) ICC manually using ITK-SNAP (spline editing capabilities, not available in Slicer). Result: Gucci-ICC.nrrd
Step 3 Register subjects to template with BRAINSFit. (Important modifications to the default parameters: --transformType Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline --initializeTransformMode CenterOfHead). Save the resulting affine and affine+bspline bulk transforms as .tfm (problems reading .mat transforms have been observed with different tools).
Step 4 Invert the transform using InvertBSpline tool, recover ICC for each subject by applying the inverse transform to the ICC of Gucci. Resampling should be performed with ApplyWarp tool from BRAINSDemonWarp package. ResampleVolume2 from Slicer does not give consistent results and has not been tested with bulk Affine+BSpline transform (bug report #709 submitted)
Step 5 Perform bias correction for each subject with N4 using the recovered ICC as the mask.
Step 6 Apply histogram matching of each ICC-masked subject to the template (Gucci) with HistogramMatchICC tool. The resulting images are skull-stripped, bias-corrected, post histogram matching.
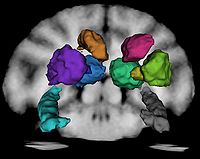
Step 7 Manually segment RIGHT SIDE subcortical structures using Gucci and the average template as the guide. This was done by Andrey under guidance of Ron. The following numbering conventions have been adopted:
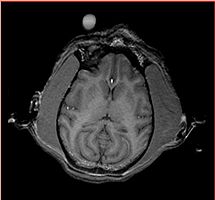
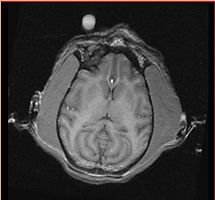
Assumption: The enhancing area on the image outside monkey head marks the RIGHT side of the subject. Label conventions: 1: CSF 2: GM 3: WM 11 and 12: Right and Left Thalamus 13 and 14: Right and Left Caudate 15 and 16: Right and Left Putamen 17 and 18: Right and Left Globus Pallidus 19 and 20: Right and Left Hippocampus
Step 8 Use ThreePointMirror extension to reflect the RIGHT SIDE labels for subcortical structures to the left side. This was done by manually placing three fiducials at the average template midline, and running the tool. The reflected LEFT SIDE labels were not in perfect alignment, and were manually edited by Andrey. Note: at this stage, these segmentations are in the original space of Gucci after discarding direction cosines from the orientation matrix.
Step 7 Align AC-PC line of the average template manually.
Step 8 Register (up to B-spline) masked, bias-corrected input subjects to the AC-PC aligned average template from the previous step. This is done with BRAINSFit tool, same parameters as in Step 3.
Step 9 Resample the axial subject images (1) with the Affine+BSpline transform and (2) with Affine transform component only, which were obtained in the previous step. Note: at this step, BSpline interpolator was used for greyscale images. Also, ICC for each subject was resampled, as well as the segmentations of subcortical structures in Gucci space. At this step, we have subcorticals in the space of the second iteration average image.
Step 10 Perform histogram matching of bias-corrected, resampled to average template space, subjects to the average template, use resampled ICC mask. The output is 10 skull-stripped subjects in the space of average template. Recompute the average templates using affinely registered and bspline registered masked scans. The result of the averaging affinely aligned subjects is the final average template, and the bspline average is the image used to segment CSF/WM/GM, and back-propagate all structures to the subjct images.
Step 11 Segment CSF,GM,WM in the bspline average using Slicer Editor module with thresholding. Approximate threshold values -- CSF: [100,7000], GM:[7000,14000], WM[1400,inf]. Use ImageLabelCombine Slicer module to merge CSF/GM/WM labels with the projected segmentations of subcortical structures. Note: subcortical segmentations overwrite 3class segmentations! UPDATE: Threshold based segmentation was substituted for semi-manual 3-class segmentation prepared by Ginger in this step of the workflow!
Step 12 Given the segmentation of subcorticals in the space of bspline registered average, use InvertBSpline tool to map subcorticals from bspline average to the space of affine average. At this step we have 3class + subcortical segmentations for all subjects affinely aligned to the initial averaged template
Step 13 Evaluate probability for each label as follows: smooth the label with ~0.5 variance Gaussian kernel, add labels from 10 subjects, divide by the number of subjects. This results in the image with intensities [0,1] for each structure.
Progress
Most recent first
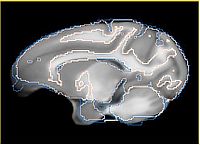
- Rhesus atlas registration improved with 2-level bspline
| Rhesus atlas registered to Calvin | ||
| Rhesus atlas seems to have a deeper sulcus, which is not recovered by registration (can it be recovered?) | ||
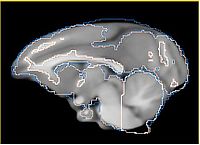
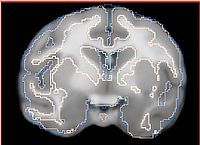
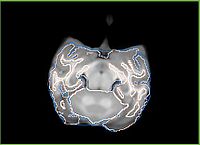
- Current effort: register rhesus atlas to vervet subjects. Preliminary results: overlay of the Gucci manual 3-class segmentation with the rhesus atlas post-affine before bspline registration (first row) and multilevel registration including bspline using BRAINSFit (second row), and another slice from the same volume, not so successful (third row).
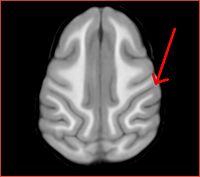
Rhesus has the frontal part bent down, and registration does not seem to capture this sufficiently. More experimentation is needed.
Also note the differences in the gyration patterns post-registration.
| (1) Overlay of vervet manual segmentation with rhesus atlas after registration up to 12 DOF (affine) | ||
| (2) Same, but after registration up to bspline transform | ||
| (3) Same registration volume as (2), but different slice | ||
- Dec 7, 2009: Initial version of the atlas ready and submitted for review to VT. v0.1: 3class segmentations were produced in the average affine template by thresholding. v0.2: 3class segmentations are resampled from semi-manual labels created by Ginger (cerebellum marked as WM). Both atlases contain: probabilistic segmentations of GM/WM/CSF + subcortical structures and average atlas image. Next steps:
- Ginger will experiment with segmentation using the new atlas, consider itkEMS (relevant study)
- Andrey will work on trying to register the UNC rhesus atlas to vervet subjects
- Nov 30, 2009:
- subcorticals now with hippocampus for one hemisphere)
- label image mirror tool in Sandbox (to simplify second hemisphere segmentation)
- Nov 23, 2009:
- Vervet Atlas Construction:Debugging bspline registration
- bspline deformation inversion tool in Sandbox (Ginger)
- Nov 19, 2009:
- ApplyWarp tool from BRAINSDemonWarp produces resampled image consistent with the output of BRAINSFit for affine+bspline bundled transform.
- N4 bias correction tested.
- ICC segmentations for subjects are ready for the subjects registered to template, bspline deformation inversion tool is in progress (Ginger)
- initial subcortical segmentations are ready (one hemisphere)
- Nov 16, 2009: Average template ready, started working on subcortical structures segmentation (AF under Ron's guidance)
- Nov 13, 2009: A significant non-rigid discrepancy between the registered image produced by BRAINSFit and the image resampled with the ResampleVolume2 and the transform produced by BRAINSFit.
- Oct 29, 2009: experiments with the input data resampled to be axis-aligned showed significant improvement of the performanc in BSplpine registration. Added a module to discard direction cosines from the input images (http://svn.na-mic.org/NAMICSandBox/trunk/VervetAtlas/Tools/DiscardDirectionCosines/). Need to re-think the workflow in light of this. Likely, we will not be able to impose AC-PC alignment on the template subject.
- Oct 27, 2009: discussed the possibility of applying groupwise registration to the vervet data with Lilla Zollei. Lilla has the data,
- Oct, 2009:
- AF: Progress with workflow stalled, since BSpline registration does not perform well with the current data
- GL: ICC prepared for Gucci, initial attempts to manually segment GM/WM. Difficulties identifying basal ganglia.
- GL: Watershed segmentation considered as a tool to help with manual segmentation. CLI module in preparation.
- AF: Experiments with EM Segmenter to segment GM/WM, ECC+CSF masked out. Not successful.
- AF: ITK 3.16 is required to handle image orientation. Slicer updated accordingly, initial problems with BRAINSFit resolved, but the results are not satisfying (TODO: put images)