Difference between revisions of "Vervet Atlas Construction:Debugging bspline registration"
| Line 108: | Line 108: | ||
===Experiment 4=== | ===Experiment 4=== | ||
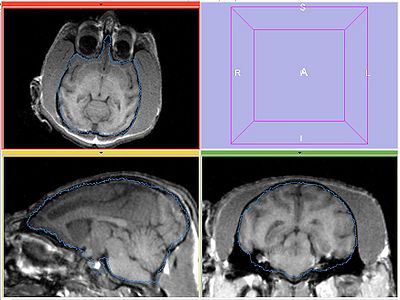
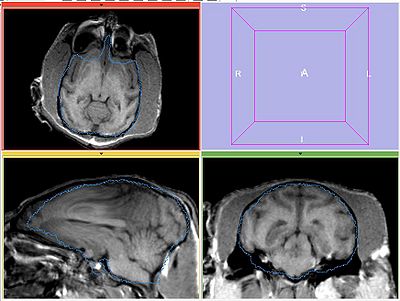
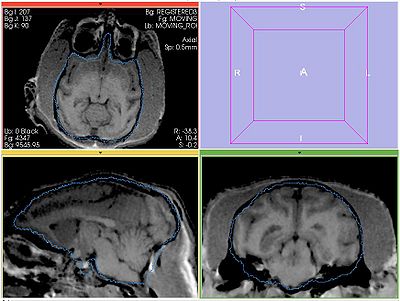
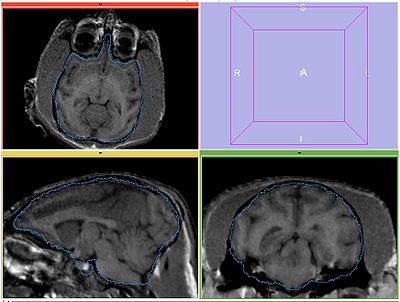
| − | Perform bias correction on the moving image prior to registration. Register MOVING_BIAS_CORRECTED/MOVING_ROI to FIXED2/FIXED_ROI. Initialize with INITIAL_TFM. | + | Perform bias correction on the moving image prior to registration. Register MOVING_BIAS_CORRECTED/MOVING_ROI to FIXED2/FIXED_ROI. Initialize with INITIAL_TFM. No histogram matching applied. |
{| | {| | ||
|[[Image:vervet_bspline_registered4.jpg|thumb|400px|REGISTERED4]] | |[[Image:vervet_bspline_registered4.jpg|thumb|400px|REGISTERED4]] | ||
|} | |} | ||
Revision as of 20:48, 24 November 2009
Home < Vervet Atlas Construction:Debugging bspline registrationContents
Introduction
Our goal is to construct probabilistic atlas of vervet monkey brain. We have 10 MR scans of vervet monkeys. Our plan is to initially select a template subject, register all subjects to the template subject. We then compute average template, and repeat the process by registering all subjects to the average template.
We use BRAINSFit Slicer plugin (latest svn checkout 327) to do all steps of registration.
Detailed description
The input includes the following items:
- FIXED1.nrrd: Fixed image1: average over subjects registered to template
- FIXED2.nrrd: Fixed image2: average with additional processing (histogram matching and bias correction), non-brain areas masked out
- MOVING.nrrd: Moving image: one of the vervet subjects
- FIXED_ROI.nrrd: brain mask for the fixed image
- MOVING_ROI.nrrd: brain mask for the moving image
- INITIAL_TFM.tfm: affine transform aligning MOVING with FIXED
- Everything in one package (together with the Slicer scene)
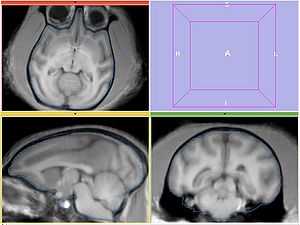
The following screeshots demonstrate good initial alignment of the images with the initial affine transformation, and the outline of ROIs. Also note that FIXED1 and FIXED2 are very similar within ROI.
Experiment1
Register MOVING/MOVING_ROI to FIXED1/FIXED_ROI. Initialize with INITIAL_TFM.
Command line:
#!/bin/bash
SLICER=/workspace/fedorov/Slicer/Release/Slicer3-build/Slicer3
BRAINSFIT=/workspace/fedorov/Slicer/Release/Slicer3-ext/BRAINSFit-build/lib/Slicer3/Plugins/BRAINSFit
FIXED_IMAGE=FIXED1.nrrd
FIXED_MASK=FIXED_ROI.nrrd
MOVING_IMAGE=MOVING.nrrd
MOVING_MASK=MOVING_ROI.nrrd
OUTPUT_IMAGE=REGISTERED1.nrrd
INIT_TFM=INITIAL_TFM.tfm
CMD="${SLICER} --launch $BRAINSFIT --fixedVolume $FIXED_IMAGE --movingVolume $MOVING_IMAGE \
--transformType BSpline --initializeTransformMode Off --initialTransform ${INIT_TFM} \
--maskInferiorCutOffFromCenter 1000 --numberOfIterations 1500 --numberOfSamples 100000 \
--minimumStepSize 0.005 --translationScale 1000 --reproportionScale 1 --skewScale 1 \
--splineGridSize 14,10,12 --outputVolumePixelType float --backgroundFillValue 0 \
--maskProcessingMode ROI --fixedVolumeTimeIndex 0 --movingVolumeTimeIndex 0 \
--medianFilterSize 0,0,0 --numberOfHistogramBins 50 --numberOfMatchPoints 10 \
--patientID ANON --studyID ANON --fixedVolumeOrigin 0,0,0 --movingVolumeOrigin 0,0,0 \
--useCachingOfBSplineWeightsMode ON --useExplicitPDFDerivativesMode AUTO --relaxationFactor 0.5 \
--maximumStepSize 0.2 --failureExitCode -1 --debugNumberOfThreads 16 --outputVolume $OUTPUT_IMAGE \
--fixedBinaryVolume $FIXED_MASK --movingBinaryVolume $MOVING_MASK"
echo $CMD
$CMD
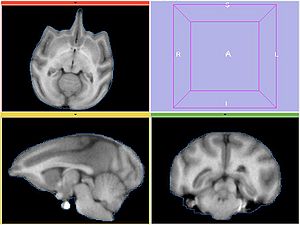
Experiment2
Register MOVING/MOVING_ROI to FIXED2/FIXED_ROI. Initialize with INITIAL_TFM. Note that the only difference from the previous experiment is the fixed image.
Command line:
#!/bin/bash
SLICER=/workspace/fedorov/Slicer/Release/Slicer3-build/Slicer3
BRAINSFIT=/workspace/fedorov/Slicer/Release/Slicer3-ext/BRAINSFit-build/lib/Slicer3/Plugins/BRAINSFit
FIXED_IMAGE=FIXED2.nrrd
FIXED_MASK=FIXED_ROI.nrrd
MOVING_IMAGE=MOVING.nrrd
MOVING_MASK=MOVING_ROI.nrrd
OUTPUT_IMAGE=REGISTERED2.nrrd
INIT_TFM=INITIAL_TFM.tfm
CMD="${SLICER} --launch $BRAINSFIT --fixedVolume $FIXED_IMAGE --movingVolume $MOVING_IMAGE \
--transformType BSpline --initializeTransformMode Off --initialTransform ${INIT_TFM} \
--maskInferiorCutOffFromCenter 1000 --numberOfIterations 1500 --numberOfSamples 100000 \
--minimumStepSize 0.005 --translationScale 1000 --reproportionScale 1 --skewScale 1 \
--splineGridSize 14,10,12 --outputVolumePixelType float --backgroundFillValue 0 \
--maskProcessingMode ROI --fixedVolumeTimeIndex 0 --movingVolumeTimeIndex 0 \
--medianFilterSize 0,0,0 --numberOfHistogramBins 50 --numberOfMatchPoints 10 \
--patientID ANON --studyID ANON --fixedVolumeOrigin 0,0,0 --movingVolumeOrigin 0,0,0 \
--useCachingOfBSplineWeightsMode ON --useExplicitPDFDerivativesMode AUTO --relaxationFactor 0.5 \
--maximumStepSize 0.2 --failureExitCode -1 --debugNumberOfThreads 16 --outputVolume $OUTPUT_IMAGE \
--fixedBinaryVolume $FIXED_MASK --movingBinaryVolume $MOVING_MASK"
echo $CMD
$CMD
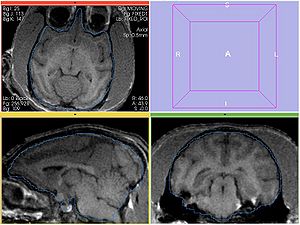
Experiment 3
Register MOVING/MOVING_ROI to FIXED2/FIXED_ROI. Initialize with INITIAL_TFM. Note that the only difference from the previous experiment is the fixed image. Per Hans' suggestion, use histogram matching (add --histogramMatching flag to BRAINSFit).
Experiment 4
Perform bias correction on the moving image prior to registration. Register MOVING_BIAS_CORRECTED/MOVING_ROI to FIXED2/FIXED_ROI. Initialize with INITIAL_TFM. No histogram matching applied.