Difference between revisions of "2009 Summer Project Week Project Segmentation of Muscoskeletal Images"
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <gallery> | + | <gallery widths="400px" perrow="6"> |
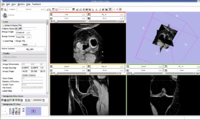
| − | Image: | + | Image:Knee.jpg| Knee MRI Image |
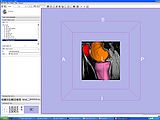
| − | Image: | + | Image:All Three.JPG|Pre-Segmented Femur/Patella/Tibia Model. |
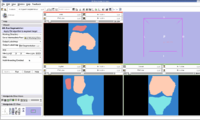
| − | Image: | + | Image:Femur Patella Tibia.jpg|EM Segmented Output. |
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Stanford: Harish Doddi, Saikat Pal, Scott Delp |
| − | * | + | * Harvard: Ron Kikinis |
| + | * Steve Pieper, Isomics, Inc. | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 15: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | The aim of this project is to develop an automatic/semi-automatic methodology to convert whole body imaging datasets into three-dimensional models for neuromuscular biomechanics and finite element simulations. | |
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 30%; float: left; padding-right: 3%;"> |
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | We focus on developing library of atlases for specific joints (eg: the knee) and evaluating EM Segmenter Algorithms. The main challenge during this process is the study of techniques for registration of a patient MRI image to a given patient MRI image (for which we already have atlas). Our approach to achieve this is to conduct parameter exploration study using the module '''Register Images Batchmake''' in Slicer 3.4 | |
| + | |||
| + | We are also investigating another technique where we try to register a given knee model to a patient MRI Image using '''Python ICP Registration'''. | ||
| + | |||
| + | Our plan for the project week is to | ||
| + | '''''a'''''. Perform and evaluate results from an extensive | ||
| + | parameter space exploration study of | ||
| + | RegisterImages Batchmake module on knee dataset. | ||
| + | '''''b'''''. Resolve issues in building Python modules from | ||
| + | slicer source code. | ||
| + | '''''c'''''. Demonstrate proof of concept on registering an | ||
| + | existing atlas (.vtk, .stl) to a target image | ||
| + | using Python ICP Registration module. | ||
| + | |||
| − | |||
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 27%; float: left;"> |
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 01:00, 29 May 2009
Home < 2009 Summer Project Week Project Segmentation of Muscoskeletal Images
Key Investigators
- Stanford: Harish Doddi, Saikat Pal, Scott Delp
- Harvard: Ron Kikinis
- Steve Pieper, Isomics, Inc.
Objective
The aim of this project is to develop an automatic/semi-automatic methodology to convert whole body imaging datasets into three-dimensional models for neuromuscular biomechanics and finite element simulations.
Approach, Plan
We focus on developing library of atlases for specific joints (eg: the knee) and evaluating EM Segmenter Algorithms. The main challenge during this process is the study of techniques for registration of a patient MRI image to a given patient MRI image (for which we already have atlas). Our approach to achieve this is to conduct parameter exploration study using the module Register Images Batchmake in Slicer 3.4
We are also investigating another technique where we try to register a given knee model to a patient MRI Image using Python ICP Registration.
Our plan for the project week is to
a. Perform and evaluate results from an extensive parameter space exploration study of RegisterImages Batchmake module on knee dataset. b. Resolve issues in building Python modules from slicer source code. c. Demonstrate proof of concept on registering an existing atlas (.vtk, .stl) to a target image using Python ICP Registration module.
Progress