Difference between revisions of "2010 Summer Project Week Liver Ablation"
From NAMIC Wiki
| (13 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
* BWH: Haiying Liu, Noby Hata, Sota Oguro | * BWH: Haiying Liu, Noby Hata, Sota Oguro | ||
* Georgetown University: Ziv Yaniv | * Georgetown University: Ziv Yaniv | ||
| + | |||
| + | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objectives</h3> | <h3>Objectives</h3> | ||
| − | * | + | * Build a system for surgical planning and navigation to guide RF liver ablation. |
| − | |||
| − | |||
</div> | </div> | ||
| Line 20: | Line 20: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | The | + | The workflow reads as follows [after segmentation]: |
| − | + | # Export the segmentation to the planning program.<br> | |
| − | + | # Run the planning program.<br> | |
| − | + | # Load the plan specified in an xml file into Slicer for visualization and modification.<br> | |
| + | # User can view the plan. Ablations are overlayed onto the image slices. Ablation locations are from the xml file and the size of the sphere is already available in Slicer before step 1.<br> | ||
| + | # User can move ablation locations.<br> | ||
| + | # Export the modified plan.<br> | ||
</div> | </div> | ||
| Line 29: | Line 32: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Got tracking data from Aurora system using IGSTK | |
| − | + | * Can read surgical plan from an xml file | |
| + | * Can write surgical plan to an xml file | ||
| + | * Working on visualization and modification of a surgical plan possibily using Measurements module | ||
</div> | </div> | ||
| Line 37: | Line 42: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | The work will be | + | The work will be evolved as a Slicer module for surgical planning. |
==References== | ==References== | ||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 13:34, 25 June 2010
Home < 2010 Summer Project Week Liver AblationKey Investigators
- BWH: Haiying Liu, Noby Hata, Sota Oguro
- Georgetown University: Ziv Yaniv
Objectives
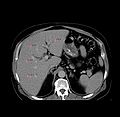
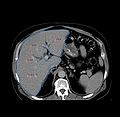
- Build a system for surgical planning and navigation to guide RF liver ablation.
Approach, Plan
The workflow reads as follows [after segmentation]:
- Export the segmentation to the planning program.
- Run the planning program.
- Load the plan specified in an xml file into Slicer for visualization and modification.
- User can view the plan. Ablations are overlayed onto the image slices. Ablation locations are from the xml file and the size of the sphere is already available in Slicer before step 1.
- User can move ablation locations.
- Export the modified plan.
Progress
- Got tracking data from Aurora system using IGSTK
- Can read surgical plan from an xml file
- Can write surgical plan to an xml file
- Working on visualization and modification of a surgical plan possibily using Measurements module
Delivery Mechanism
The work will be evolved as a Slicer module for surgical planning.