2012 Winter Project Week:CTLiverRegistration
From NAMIC Wiki
Home < 2012 Winter Project Week:CTLiverRegistration
Contents
Key Investigators
- AZE Research and Development, Karl Diedrich
- Brigham and Women's Hospital, Nobuhiko Hata
- Brigham and Women's Hospital, Atsushi Yamada
Objective
Register CT images of livers taken at different time points for the purpose of detecting changes in liver tumors.
Approach, Plan
- Acquire unregistered 4D liver data sets.
- Segment bone using upper and lower Hounsfield unit threshold.
- Create a distance map from the threshold segmented bones. Look into using vtkITKDistanceTransform for distance map. The bones are very sparse making poor registration subjects.
- Conduct a fixed registration of the fixed and moving bones near the liver using BRAINSfit in slicer.
- Apply the resulting transform to the full moving liver data set.
Progress
- Acquired additional unregistered 4D liver time series data sets for development and testing from http://midas.kitware.com/community/view/47.
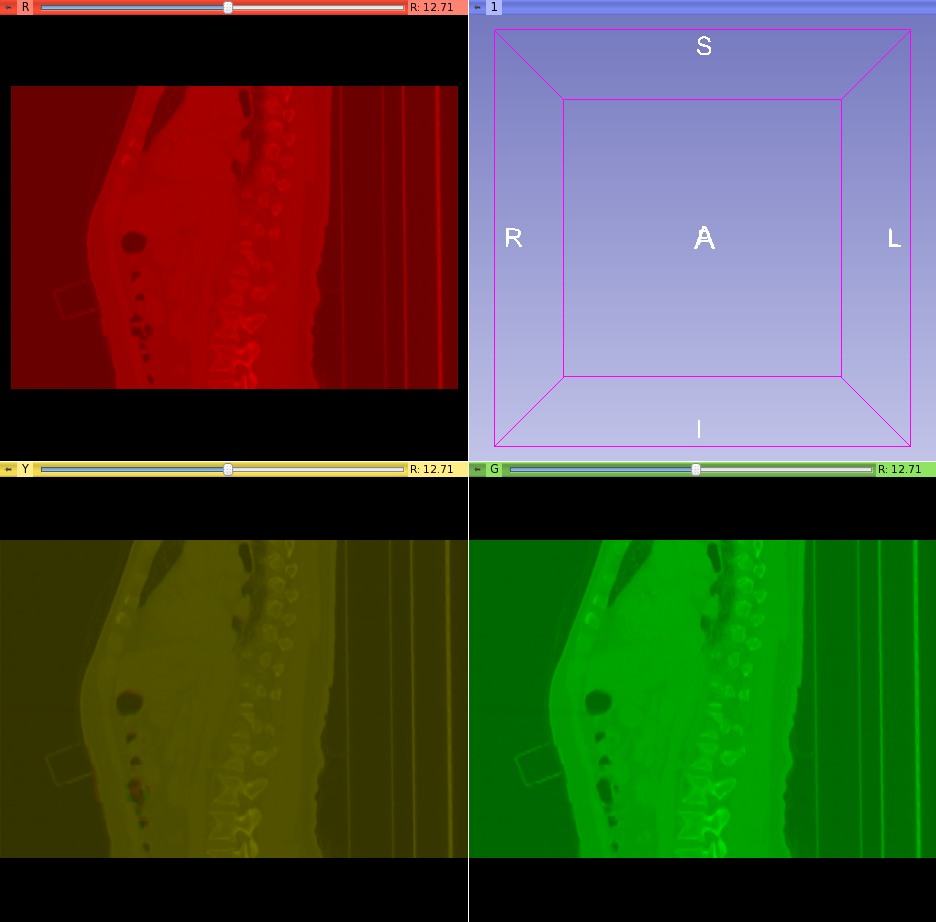
- Used Slicer to segment the bones using a lower threshold of 252. Segmented the bones with a label of 255 to allow blurring. See upper left images below.
- ITK contains a filter class, itkSignedDanielssonDistanceMapImageFilter, http://www.itk.org/Doxygen/html/classitk_1_1SignedDanielssonDistanceMapImageFilter.html for creating a distance map. Look into using vtkITKDistanceTransform for distance map. Need to develop a Slicer CLI module for generating distance maps of the fixed and moving bone images.
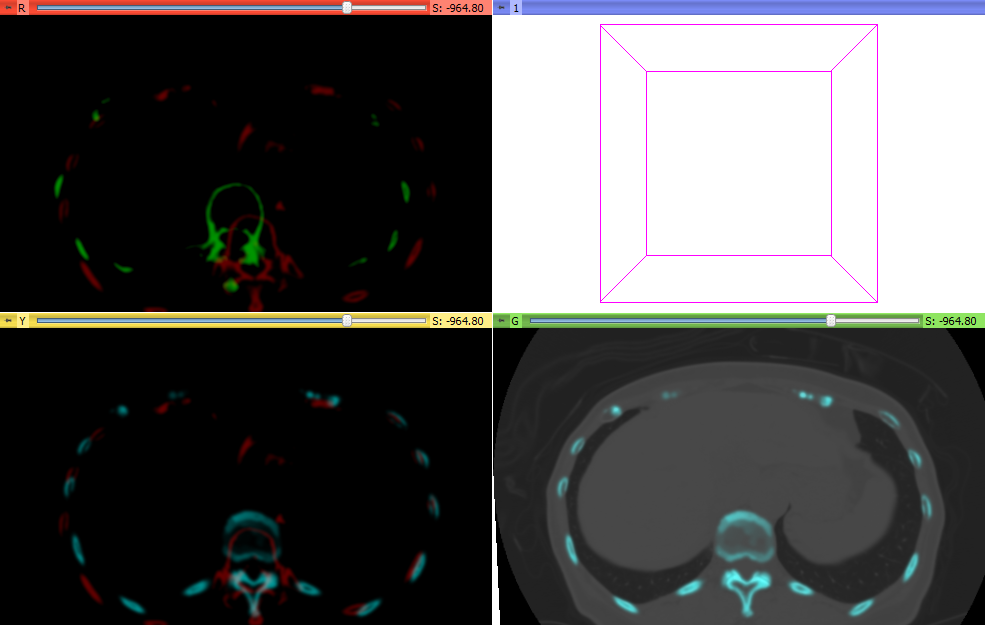
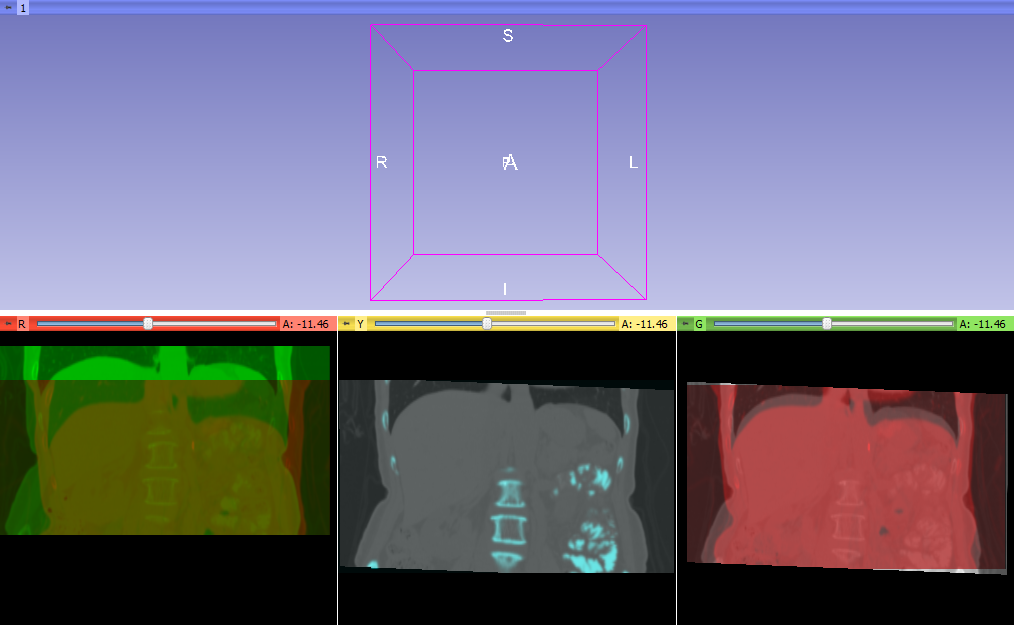
- Registered two time points of the same patient as fixed and moving images in BRAINSfit using Initialize Transform Mode: useGeometryAlign Registration Phase: Rigid (6 DOF) AND Rigid+Scale (7DOF) AND Rigid+Scale+Skew (10 DOF). See lower left images below.
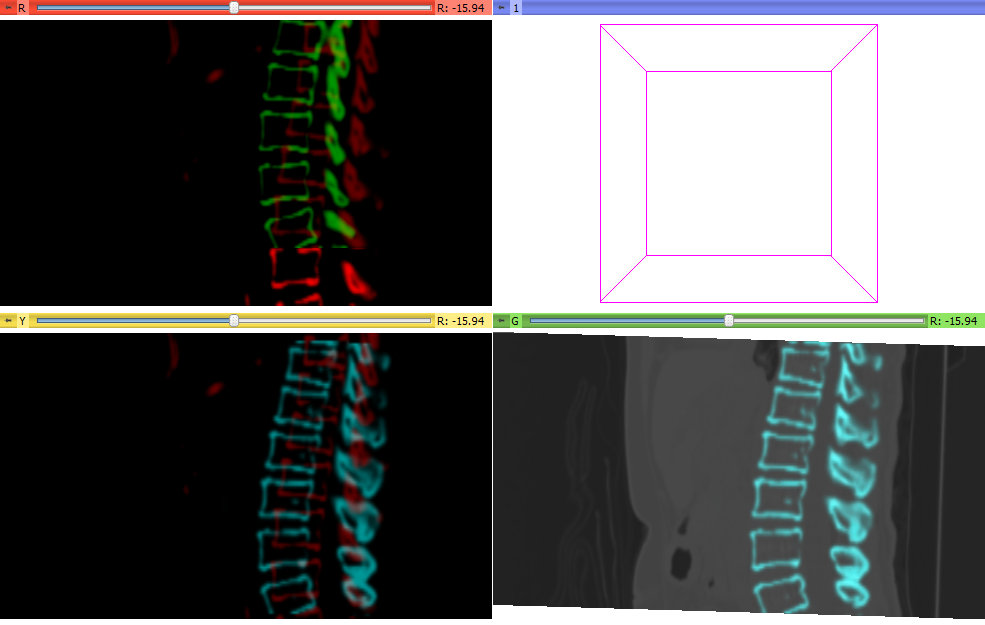
- Applied the resulting transform to the full moving liver data set in the Slicer Data module Nodes panel by dragging the moving liver data set onto the output transform name. See lower right images below.
Delivery mechanism
Develop a Slicer CLI module using the ITK distance map itkSignedDanielssonDistanceMapImageFilter, http://www.itk.org/Doxygen/html/classitk_1_1SignedDanielssonDistanceMapImageFilter.html. Then chain the steps together in Python.
References
Hans Johnson, Greg Harris, Kent Williams. BRAINSFit: Mutual Information Rigid Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. The Insight Journal. 2009.