2014 Project Week:CardiacCongenitalSegmentation
From NAMIC Wiki
Home < 2014 Project Week:CardiacCongenitalSegmentation
Key Investigators
Danielle Pace, MIT
Polina Golland, MIT
Project Description
Objective
- Develop a semi-automatic segmentation algorithm for cardiac MR images of patients with congenital heart defects.
- Goal is to build a surface model showing the endocardial and epicardial boundaries, for surgical planning.
- Challenges:
- Very large inter-subject variability due to heart defects
- Intensity inhomogeneities within myocardium and blood pool
- Similar intensity distributions within adjacent tissues (e.g. liver, chest muscle)
Approach, Plan
- Try existing open-source tools for segmentation and registration on the five datasets that we have so far.
- See where these methods fail, to focus our efforts for developing algorithms for segmenting hearts with congenital defects.
Progress
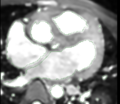
- Focused on blood pool segmentation in one test case. Tried:
- CARMA tools: isolated connected / connected threshold operators
- Editor level tracing effect
- Editor Fast Marching effect
- Editor Grow Cuts effect
- Carrera interactive segmentation
- Robust statistic active contour segmentation
- Best tool = Carrera interactive segmentation
- Main difficulty = small chamber/vessel walls assigned as blood pool, but these can be fixed within Carrera somewhat easily
- Also tried affine registrations across my 5 subjects, using BRAINSFIT
- Works very roughly, as expected
- Next steps:
- Try Carrera on additional datasets for blood pool segmentation
- Myocardium segmentation is still a challenge
- Thanks to: Josh Cates, Salma Bengali, Yi Gao, Ivan Kolesov for your help and suggestions!