Difference between revisions of "DBP1:Harvard"
| (30 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Back to [[DBP1:Main|NA-MIC DBP 1]] | Back to [[DBP1:Main|NA-MIC DBP 1]] | ||
__NOTOC__ | __NOTOC__ | ||
| − | |||
= Overview of Harvard PNL DBP 1 = | = Overview of Harvard PNL DBP 1 = | ||
| − | + | The Harvard Driving Biological Project uses structural MRI, diffusion-weighted MRI, and fMRI to study the neural bases of schizophrenia and related psychiatric disorders. The DBP is centered in the Psychiatry Neuroimaging Laboratory (PNL) at Brigham and Women's Hospital in Boston, MA. | |
| + | |||
| + | For more information about the PNL, please visit the [http://pnl.bwh.harvard.edu| lab website.] | ||
| + | |||
= Harvard PNL Projects = | = Harvard PNL Projects = | ||
| − | {| cellpadding="10" | + | {| cellpadding="10" style="text-align:left;" |
| − | | style="width:15%" | [[Image: | + | | style="width:15%" | [[Image:Shapecaudate.png|200px]] |
| style="width:85%" | | | style="width:85%" | | ||
| − | == [[ | + | == [[Projects:ShapeAnalysisOfBrainStructures|Shape Analysis of Brain Structures]] == |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | In this work, we present shape analysis algorithms for brain structures. [[Projects:ShapeAnalysisOfBrainStructures|More...]] | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> G Gerig, S Joshi, T Fletcher, K Gorczowski, S Xu, S M. Pizer, M Styner: Statistics of populations of images and its embedded objects: Driving applications in neuroimaging, IEEE Symposium on Biomedical Imaging ISBI 2006. |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:CingulumAllSubjectsFibers.png|200px]] |
| | | | | | ||
| − | == [[ | + | == [[Projects:DTIClustering|DTI Fiber Clustering and Fiber-Based Analysis]] == |
| − | + | The goal of this project is to provide structural description of the white matter architecture as a partition into coherent fiber bundles and clusters, and to use these bundles for quantitative measurement. [[Projects:DTIClustering|More...]] | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New:'''</font> Monica E. Lemmond, Lauren J. O'Donnell, Stephen Whalen, Alexandra J. Golby. Characterizing Diffusion Along White Matter Tracts Affected by Primary Brain Tumors. Accepted to HBM 2007. |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Progress_Registration_Segmentation_Shape.jpg|200px]] |
| − | | | | + | | | |
| − | == [[ | + | == [[Projects:ShapeBasedSegmentationAndRegistration|Shape Based Segmentation and Registration]] == |
| − | + | This type of algorithm assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. [[Projects:ShapeBasedSegmentationAndRegistration|More...]] | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Medical Image Analysis,11(6), pp. 465-477, 2007. <b>Best Paper Award MICCAI 2006 </b> |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:Striatum1.png|200px|]] |
| | | | | | ||
| − | == [[ | + | == [[Projects:RuleBasedStriatumSegmentation|Rule-Based Striatum Segmentation]] == |
| − | + | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. [[Projects:RuleBasedStriatumSegmentation|More...]] | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Al-Hakim, et al. Parcellation of the Striatum. SPIE MI 2007. |
|- | |- | ||
| − | | | [[ | + | | | [[Image:DTIregistration200.png|200px]] |
| | | | | | ||
| − | == [[ | + | == [[Projects:DTIProcessingTools|Diffusion Tensor Image Processing Tools]] == |
| − | + | We implement the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al. Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction. | |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> We have recently developed software for eddy current correction. |
|- | |- | ||
| − | | | | + | | | |
| | | | | | ||
== [[DBP:Harvard:Collaboration:Slicer|Engineering: Slicer Improvement and Testing]] == | == [[DBP:Harvard:Collaboration:Slicer|Engineering: Slicer Improvement and Testing]] == | ||
| − | + | This ongoing work conducts testing and development of new features in Slicer and provides a bridge between developers and researchers using the software. [[DBP:Harvard:Collaboration:Slicer|More...]] | |
| − | |||
| − | |||
|- | |- | ||
| − | | | [[ | + | | | [[Image:Trainingslide.png|200px]] |
| | | | | | ||
== [[DBP:Harvard:Collaboration:Training|Training: Training Material and Expert Users Feedback]] == | == [[DBP:Harvard:Collaboration:Training|Training: Training Material and Expert Users Feedback]] == | ||
| − | + | Work in this area has created training materials for Slicer software with the input of expert users. [[DBP:Harvard:Collaboration:Training|More...]] | |
| − | |||
| − | |||
|- | |- | ||
| − | | | | + | | | |
| | | | | | ||
== [[DBP:Harvard:Collaboration:Toronto|Biology: Genetics and Imaging (Toronto)]] == | == [[DBP:Harvard:Collaboration:Toronto|Biology: Genetics and Imaging (Toronto)]] == | ||
| − | + | This collaboration is centered around a study of psychosis in schizophrenia and bipolar disorder, using a combination of genetics and diffusion tensor imaging. [[DBP:Harvard:Collaboration:Toronto|More...]] | |
| − | + | |} | |
| − | | | + | For more introductory information, follow this [[DBP:Harvard|link]]. |
Latest revision as of 19:06, 11 July 2009
Home < DBP1:HarvardBack to NA-MIC DBP 1
Overview of Harvard PNL DBP 1
The Harvard Driving Biological Project uses structural MRI, diffusion-weighted MRI, and fMRI to study the neural bases of schizophrenia and related psychiatric disorders. The DBP is centered in the Psychiatry Neuroimaging Laboratory (PNL) at Brigham and Women's Hospital in Boston, MA.
For more information about the PNL, please visit the lab website.
Harvard PNL Projects
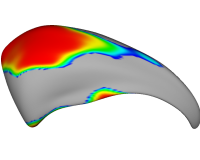
|
Shape Analysis of Brain StructuresIn this work, we present shape analysis algorithms for brain structures. More... New: G Gerig, S Joshi, T Fletcher, K Gorczowski, S Xu, S M. Pizer, M Styner: Statistics of populations of images and its embedded objects: Driving applications in neuroimaging, IEEE Symposium on Biomedical Imaging ISBI 2006. |
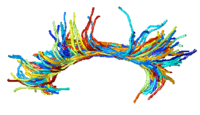
|
DTI Fiber Clustering and Fiber-Based AnalysisThe goal of this project is to provide structural description of the white matter architecture as a partition into coherent fiber bundles and clusters, and to use these bundles for quantitative measurement. More... New: Monica E. Lemmond, Lauren J. O'Donnell, Stephen Whalen, Alexandra J. Golby. Characterizing Diffusion Along White Matter Tracts Affected by Primary Brain Tumors. Accepted to HBM 2007. |
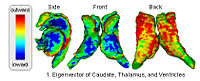
|
Shape Based Segmentation and RegistrationThis type of algorithm assigns a tissue type to each voxel in the volume. Incorporating prior shape information biases the label assignment towards contiguous regions that are consistent with the shape model. More... New: K.M. Pohl, J. Fisher, S. Bouix, M. Shenton, R. W. McCarley, W.E.L. Grimson, R. Kikinis, and W.M. Wells. Using the Logarithm of Odds to Define a Vector Space on Probabilistic Atlases. Medical Image Analysis,11(6), pp. 465-477, 2007. Best Paper Award MICCAI 2006 |
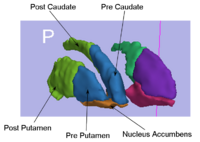
|
Rule-Based Striatum SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. More... New: Al-Hakim, et al. Parcellation of the Striatum. SPIE MI 2007. |
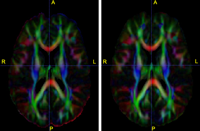
|
Diffusion Tensor Image Processing ToolsWe implement the diffusion weighted image (DWI) registration model from the paper of G.K.Rohde et al. Patient head motion and eddy currents distortion cause artifacts in maps of diffusion parameters computer from DWI. This model corrects these two distortions at the same time including brightness correction. New: We have recently developed software for eddy current correction. |
Engineering: Slicer Improvement and TestingThis ongoing work conducts testing and development of new features in Slicer and provides a bridge between developers and researchers using the software. More... | |
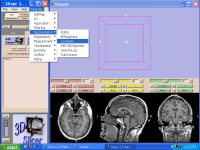
|
Training: Training Material and Expert Users FeedbackWork in this area has created training materials for Slicer software with the input of expert users. More... |
Biology: Genetics and Imaging (Toronto)This collaboration is centered around a study of psychosis in schizophrenia and bipolar disorder, using a combination of genetics and diffusion tensor imaging. More... |
For more introductory information, follow this link.