Difference between revisions of "Projects:GroupwiseRegistration"
| Line 77: | Line 77: | ||
''Progress'' | ''Progress'' | ||
| + | |||
| + | We tested the groupwise registration algorithm on a MR brain dataset and | ||
| + | compared the results to a pairwise approach \cite{joshi}. | ||
| + | The dataset consists of 50 MR brain images of three subgroups: | ||
| + | schizophrenics, affected disorder and normal control patients. | ||
| + | MR images are T1 scans with $256\times256\times128$ voxels | ||
| + | and $0.9375\times0.9375\times1.5$~$\mbox{mm}^3$ spacing. | ||
| + | MR images are preprocessed by skull stripping. | ||
| + | To account for global intensity variations, images are normalized | ||
| + | to have zero mean and unit variance. | ||
| + | For each image in the dataset, an automatic tissue classification \cite{pohl} | ||
| + | was performed, yielding gray matter (GM), white matter (WM) and cerebro-spinal | ||
| + | fluid (CSF) labels. In addition, manual segmentations of four subcortical regions | ||
| + | (left and right hippocampus and amygdala) and four cortical regions (left and right superior temporal | ||
| + | gyrus and para-hippocampus) were available for each MR image. | ||
= Key Investigators = | = Key Investigators = | ||
Revision as of 20:16, 9 November 2007
Home < Projects:GroupwiseRegistrationBack to NA-MIC_Collaborations, MIT Algorithms
Non-rigid Groupwise Registration
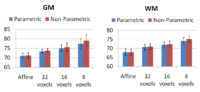
In this work, we extend a previously demonstrated entropy based groupwise registration method to include a free-form deformation model based on B-splines. We provide an efficient implementation using stochastic gradient descents in a multi-resolution setting. We demonstrate the method in application to a set of 50 MRI brain scans and compare the results to a pairwise approach using segmentation labels to evaluate the quality of alignment. Our results indicate that increasing the complexity of the deformation model improves registration accuracy significantly, especially at cortical regions.
Description
Objective Function
In order to align all subjects in the population,
we consider sum of pixelwise entropies as a joint alignment criterion.
The justification for this approach is that if the images are aligned properly,
intensity values at corresponding coordinate locations from all the images
will form a low entropy distribution.
This approach does not require the use of a reference subject; all
subjects are simultenously driven to the common tendency of the population.
Implementation
We provide an efficient optimization scheme by using line search with the gradient descent algorithm. For computational efficiency, we employ a stochastic subsampling procedure \cite{pluim}. In each iteration of the algorithm, a random subset is drawn from all samples and the objective function is evaluated only on this sample set. The number of samples to be used in this method depends on the number of the parameters of the deformation field. Using the number of samples on the order of the number of variables works well in practice.
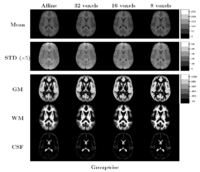
To obtain a dense deformation field capturing anatomical variations at different scales, we gradually increase the complexity of the deformation field by refining the grid of B-spline control points. First, we perform a global registration with affine transforms. Then, we use affine transforms to initialize a low resolution deformation field at a grid spacing around $32$~voxels. We increase the resolution of the deformation field to $8$~voxels by using registration results at coarser grids to initialize finer grids.
As in every iterative search algorithm, local minima pose a significant problem. To avoid local minima we use a multi-resolution optimization scheme for each resolution level of the deformation field. The registration is first performed at a coarse scale by downsampling the input. Results from coarser scales are used to initialize optimization at finer scales. For each resolution level of the deformation field we used a multi-resolution scheme of three image resolution levels.
We implemented our groupwise registration method in a multi-threaded fashion using Insight Toolkit (ITK) and made the implementation publicly available \cite{itk}. We run experiments using a dataset of 50 MR images with $256\times256\times128$ voxels on a workstation with four CPUs and 8GB of memory. The running time of the algorithm is about 20 minutes for affine registration and two days for non-rigid registration of the entire dataset. The memory requirement of the algorithm depends linearly on the number of input images and was around 3GB for our dataset.
Progress
We tested the groupwise registration algorithm on a MR brain dataset and compared the results to a pairwise approach \cite{joshi}. The dataset consists of 50 MR brain images of three subgroups: schizophrenics, affected disorder and normal control patients. MR images are T1 scans with $256\times256\times128$ voxels and $0.9375\times0.9375\times1.5$~$\mbox{mm}^3$ spacing. MR images are preprocessed by skull stripping. To account for global intensity variations, images are normalized to have zero mean and unit variance. For each image in the dataset, an automatic tissue classification \cite{pohl} was performed, yielding gray matter (GM), white matter (WM) and cerebro-spinal fluid (CSF) labels. In addition, manual segmentations of four subcortical regions (left and right hippocampus and amygdala) and four cortical regions (left and right superior temporal gyrus and para-hippocampus) were available for each MR image.
Key Investigators
- MIT: Serdar K Balci, Polina Golland, Sandy Wells
Publications
- S.K. Balci, P. Golland, M.E. Shenton, W.M. Wells III. Free-Form B-spline Deformation Model for Groupwise Registration. In Proceedings of MICCAI 2007 Statistical Registration Workshop: Pair-wise and Group-wise Alignment and Atlas Formation, 23-30, 2007.