Difference between revisions of "DBP2:UNC:Regional Cortical Thickness Pipeline"
| Line 95: | Line 95: | ||
|} | |} | ||
</center> | </center> | ||
| + | |||
| + | === Done === | ||
| + | :* Workflow for individual analysis (Slicer3 external module using BatchMake) | ||
| + | :* 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC" | ||
=== In progress === | === In progress === | ||
| − | :* | + | :* Pediatric atlases (T1-weighted image, parcellation map, probability maps) available to the community (MIDAS : NAMIC stuff) |
| − | : | ||
:* Regional analysis to compare our method with results from FreeSurfer | :* Regional analysis to compare our method with results from FreeSurfer | ||
| − | :* | + | :* ARCTIC executables (UNC external modules for Slicer3D), Source code (SVN) and Tutorial dataset available on NITRC |
=== Future work === | === Future work === | ||
| − | :* | + | :* Workflow for group analysis (KWWidgets application using BatchMake) |
| − | |||
Revision as of 01:16, 11 December 2008
Home < DBP2:UNC:Regional Cortical Thickness PipelineBack to UNC Cortical Thickness Roadmap
Objective
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness : ARCTIC (Automatic Regional Cortical ThICkness)
General information
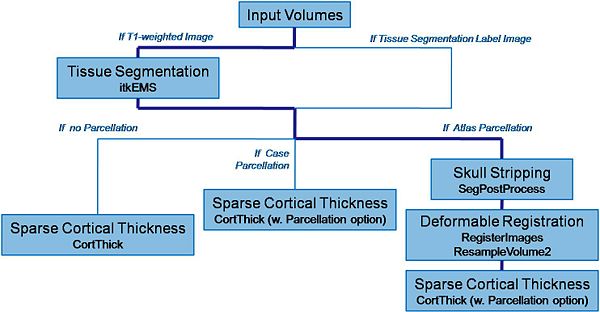
Main pipeline description (steps)
Input: RAW images (T1-weighted, T2-weighted, PD-weighted images)
- 1. Tissue segmentation
- Tool: itkEMS (UNC Slicer3 external module)
- 2. Skull stripping
- Tool: SegPostProcess (UNC Slicer3 external module)
- 3. Deformable registration of pediatric regional atlas
- 3.1 Deformable registration of T1-weighted pediatric atlas
- Tool: RegisterImages (Slicer3 module)
- 3.2. Applying transformation to the parcellation map
- Tool: ResampleVolume2 (Slicer3 module)
- 3.1 Deformable registration of T1-weighted pediatric atlas
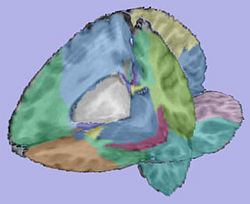
- 4. Cortical Thickness
- Tool: CortThick (UNC Slicer3 module)
- 1. Tissue segmentation
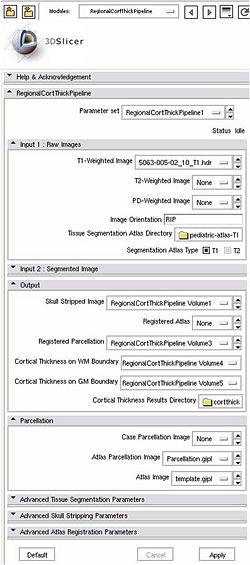
All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. The user can thus compute an individual regional cortical thickness analysis by running the 'RegionalCortThickPipeline' module, either within Slicer3 or as a command line.
Usage (Command Line)
Inputs: T1-weighted image, T1-weigthed atlas, regional atlas (parcellation map)
0. Before using the pipeline
Add the executables in the PATH.
-tcsh usage : setenv PATH ARCTIC-Executables-Directory/:Slicer3D-Plugins-Directory/:Batchmake-Drectory/:${PATH}
-bash usage : export PATH=ARCTIC-Executables-Directory/:Slicer3D-Plugins-Directory/:Batchmake-Drectory/:${PATH}
Set the pipeline environment variable
-tcsh usage : setenv BatchmakeWrapper_Dir Batchmake-Wrapper-Directory/ -bash usage : export BatchmakeWrapper_Dir=Batchmake-Wrapper-Directory/
Set the pipeline as a Slicer3D module
Within Slicer3D : View->Applications Settings->Add a preset and then select the ARCTIC-Executables-Directory/
A. Pipeline command line
RegionalCortThickPipeline --T1 Image_T1.gipl --segAtlasDir TissueSegmentationAtlasDirectory/ --atlas Atlas.gipl --atlasParcellation Parcellation.gipl --SaveWM WMCorticalThicknessMap --SaveGM GMCorticalThicknessMap
B. Step by step command line
- 1. Tissue segmentation
- Input: EMSparam.xml
- Output: Image_Corrected_EMS.gipl, Label.gipl
- 1. Tissue segmentation
itkEMSCLP --XMLFile EMSparam.xml
- 2. Skull stripping
- Input: Label.gipl, Image_Corrected_EMS.gipl
- Output: Image_Corrected_EMS_Stripped.gipl, BinaryMask.gipl (optional)
- 2. Skull stripping
SegPostProcessCLP Label.gipl Image_Corrected_EMS_Stripped.gipl --skullstripping Image_Corrected_EMS.gipl
- 3. Deformable registration of pediatric regional atlas
- 3.1 Deformable registration of T1-weighted pediatric atlas
- Input: Atlas.gipl, Image_Corrected_EMS_Stripped.gipl
- Output: Atlas_Registered.gipl, Atlas_Registered_Transform.txt
- 3.1 Deformable registration of T1-weighted pediatric atlas
- 3. Deformable registration of pediatric regional atlas
RegisterImages Image_Corrected_EMS_Stripped.gipl Atlas.gipl –resampledImage Atlas_Registered.gipl –saveTransform Atlas_Registered_Transform.txt –registration PipelineBSpline
- 3.2. Applying transformation to the parcellation map
- Input: Parcellation.gipl, Atlas_Registered_Transform.txt, Image_Corrected_EMS_Stripped.gipl
- Output: Parcellation_Registered.gipl
- 3.2. Applying transformation to the parcellation map
ResampleVolume2 Parcellation.gipl Parcellation_Registered.gipl -f Atlas_Registered_Transform.txt -i nn -R Image_Corrected_EMS_Stripped.gipl
- 4. Cortical Thickness
- Input: Parcellation_Registered.gipl, Label.gipl
- Output: CortThick_Result_Dir/, WMCorticalThicknessMap, GMCorticalThicknessMap
- 4. Cortical Thickness
CortThickCLP CortThick_Result_Dir/ --par Parcellation_Registered.gipl --inputSeg Label.gipl --SaveWM WMCorticalThicknessMap --SaveGM GMCorticalThicknessMap
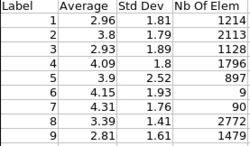
Analysis on a small pediatric dataset
Tests have been computed on a small pediatric dataset, including 2 years old and 4 years old cases
- 16 Autistic cases
- 1 developmental delay
- 3 normal control
We would like to perform a regional analysis (Pearson correlation analysis) to compare our method with FreeSurfer.
Done
- Workflow for individual analysis (Slicer3 external module using BatchMake)
- 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC"
In progress
- Pediatric atlases (T1-weighted image, parcellation map, probability maps) available to the community (MIDAS : NAMIC stuff)
- Regional analysis to compare our method with results from FreeSurfer
- ARCTIC executables (UNC external modules for Slicer3D), Source code (SVN) and Tutorial dataset available on NITRC
Future work
- Workflow for group analysis (KWWidgets application using BatchMake)