Difference between revisions of "Meningioma growth modeling with TumorSim"
| Line 7: | Line 7: | ||
'''NOTE: red outlines the boundary of the label corresponding to the initial tumor seed!''' | '''NOTE: red outlines the boundary of the label corresponding to the initial tumor seed!''' | ||
| − | <flash>file=Seed_1_simulation.swf|width= | + | <flash>file=Seed_1_simulation.swf|width=500|height=400|quality=best</flash> |
==Seed 3 (ventricle)== | ==Seed 3 (ventricle)== | ||
| − | <flash>file=Seed_3_simulation.swf|width= | + | <flash>file=Seed_3_simulation.swf|width=500|height=400|quality=best</flash> |
=tumorsim1.1.rc4 experiments= | =tumorsim1.1.rc4 experiments= | ||
Revision as of 15:25, 2 October 2009
Home < Meningioma growth modeling with TumorSimThis is a follow-up project for Meningioma growth simulation project within 2009 Boston programming week project.
Contents
tumorsim1.1rc2 experiments
Seed 1 (default)
NOTE: red outlines the boundary of the label corresponding to the initial tumor seed!
<flash>file=Seed_1_simulation.swf|width=500|height=400|quality=best</flash>
Seed 3 (ventricle)
<flash>file=Seed_3_simulation.swf|width=500|height=400|quality=best</flash>
tumorsim1.1.rc4 experiments
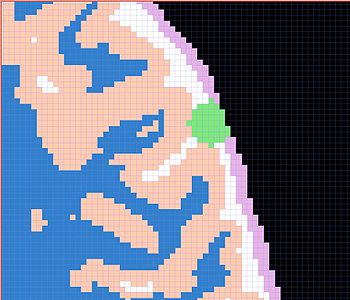
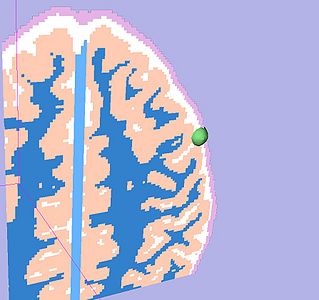
| All the following experiments are based on the "thin dura" input labels, and the tumor seed image shown on the right. The tumor label is shown in green color. Note that the tumor is touching the gray matter label. |
Also, all of the experiments use the following configuration file, where POISSON value is substituted with 0.5, 0.4, and 0.2, with the simulation results shown below.
<?xml version="1.0"?> <tumor-simulation-parameters> <dataset-name>SimMeningioma001</dataset-name> <input-directory>../TumorSimInputWithThinDura</input-directory> <output-directory>./0.05</output-directory> <brain-young-modulus>694</brain-young-modulus> <brain-poisson-ratio>POISSON</brain-poisson-ratio> <deformation-seed>../SlicerScenes/NewSeed3-1.nrrd</deformation-seed> <deformation-iterations>10</deformation-iterations> <deformation-initial-pressure>3.0</deformation-initial-pressure> <deformation-kappa>1e+06</deformation-kappa> <deformation-damping>1</deformation-damping> <deformation-solver-iterations>8</deformation-solver-iterations> <infiltration-solver-iterations>2</infiltration-solver-iterations> <infiltration-iterations>2</infiltration-iterations> <infiltration-time-step>1e-6</infiltration-time-step> <infiltration-early-time>0.5e-6</infiltration-early-time> <infiltration-body-force-iterations>2</infiltration-body-force-iterations> <infiltration-body-force-coefficient>10.0</infiltration-body-force-coefficient> <contrast-enhancement-type>uniform</contrast-enhancement-type> <number-of-threads>16</number-of-threads>
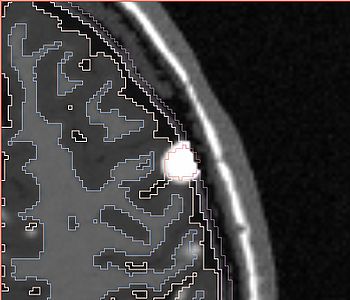
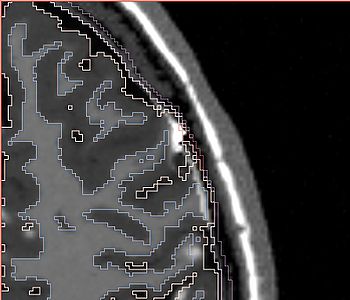
The following screenshots capture the same axial slice (#119) of the following images in order: (1) image containing the tumor probabilities, and (2) the simulated Gad enhanced image, both overlayed with the warped tumor/brain tissues labels.
| Poisson ratio 0.5
(initial value) |
| Poisson ratio 0.4 |
| Poisson ratio 0.2 |
Standing questions/items
- limited tumor growth/surrounding brain deformation with 10 iterations -- try more iterations?
- reduction of Poisson ratio makes results very unrealistic