Difference between revisions of "Projects:PathologyAnalysis"
(Created page with 'Back to Utah 2 Algorithms __NOTOC__ = Lesion Segmentation = = Description = Quantification, analysis and display of brain pathology such as white matter le…') |
|||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| − | = | + | = Analysis of Brain Images with Pathological Changes = |
= Description = | = Description = | ||
| − | + | Traumatic brain injury (TBI) occurs when an external force traumatically injures the brain. TBI is a major cause of death and disability worldwide, especially in children and young adults. TBI affects 1.4 million Americans annually. The UCLA medical school has been working on this topic for years. | |
| + | |||
| + | On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury. | ||
| + | |||
| + | We will work on an extension of ABC for TBI datasets with the clinical goal to investigate alterations in cortical thickness, subsequent ventricular, and white matter changes in patients with TBI. | ||
| + | |||
<center> | <center> | ||
Revision as of 14:24, 23 March 2011
Home < Projects:PathologyAnalysisBack to Utah 2 Algorithms
Analysis of Brain Images with Pathological Changes
Description
Traumatic brain injury (TBI) occurs when an external force traumatically injures the brain. TBI is a major cause of death and disability worldwide, especially in children and young adults. TBI affects 1.4 million Americans annually. The UCLA medical school has been working on this topic for years.
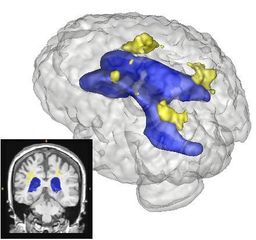
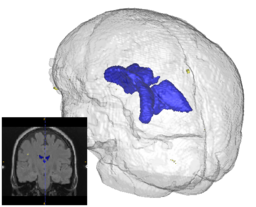
On anatomical MRI scans, to quantitatively analyze the cortical thickness, white matter changes, we need to have a good segmentation on TBI images. However, for TBI data, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury.
We will work on an extension of ABC for TBI datasets with the clinical goal to investigate alterations in cortical thickness, subsequent ventricular, and white matter changes in patients with TBI.
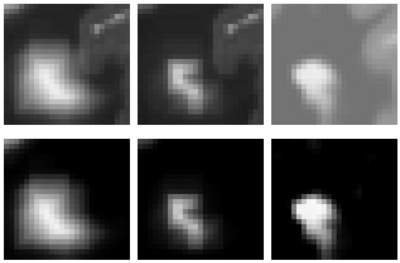
In addition to the identification of the location and shape of lesions in 3D, we are interested in analyzing the longitudinal series of brain MRI showing lesions. For this purpose, we have developed a method for estimating a physical model for lesion formation. The model that we use is an approximation using a reaction-diffusion process that is based on expected diffusion properties (as observed through DTI). This approach gives a richer parametrization of lesion changes in addition to volume and location, as the model estimation provides descriptions of growth and spread for individual lesions. In the future, we plan to incorporate this approach for analyzing lesion MRI of a subject over time by characterizing the change patterns through the physical model parameters.
Key Investigators
- Utah Algorithms: Marcel Prastawa, Guido Gerig
- Clinical Collaborators
- MIND: Jeremy Bockholt, Mark Scully
Publications
In Print