Difference between revisions of "DBP2:UNC:Regional Cortical Thickness Pipeline"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (70 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[DBP2:UNC:Cortical_Thickness_Roadmap | UNC Cortical Thickness Roadmap]] | + | Back to [[DBP2:UNC:Cortical_Thickness_Roadmap | UNC Cortical Thickness Roadmap]] |
| − | + | ||
| + | [[Image:ArcticLogo.png|thumb|400px|ARCTIC Logo|right]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Objective == | == Objective == | ||
| Line 13: | Line 8: | ||
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness. | We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness. | ||
| − | This page describes the related pipeline with its basic components. | + | This page describes the related pipeline with its basic components, as well as its validation. |
| + | |||
| − | == | + | == Pipeline overview == |
| − | + | A Slicer3 high-level module for individual cortical thickness analysis has been developed: ARCTIC (Automatic Regional Cortical ThICkness) | |
Input: RAW images (T1-weighted, T2-weighted, PD-weighted images) | Input: RAW images (T1-weighted, T2-weighted, PD-weighted images) | ||
:* '''1. Tissue segmentation''' | :* '''1. Tissue segmentation''' | ||
| + | :** Probabilistic atlas-based automatic tissue segmentation via an Expectation-Maximization scheme | ||
:** Tool: itkEMS (UNC Slicer3 external module) | :** Tool: itkEMS (UNC Slicer3 external module) | ||
| − | :* '''2. | + | :* '''2. Regional atlas deformable registration''' |
| − | :** Tool: SegPostProcess (UNC Slicer3 external module) | + | :** 2.1 Skull stripping using previously computed tissue segmentation label image |
| − | :* | + | :*** Tool: SegPostProcess (UNC Slicer3 external module) |
| − | :** | + | :** 2.2 T1-weighted atlas deformable registration |
| − | :***Tool: RegisterImages (Slicer3 module) | + | :*** B-spline pipeline registration |
| − | :** 3 | + | :*** Tool: RegisterImages (Slicer3 module) |
| + | :** 2.3. Applying transformation to the parcellation map | ||
:*** Tool: ResampleVolume2 (Slicer3 module) | :*** Tool: ResampleVolume2 (Slicer3 module) | ||
| − | :* ''' | + | :* '''3. Cortical thickness measurement''' |
| − | :** Tool: CortThick (UNC Slicer3 module) | + | :** Sparse asymmetric local cortical thickness |
| + | :** Tool: CortThick (UNC Slicer3 external module) | ||
| + | :* '''4. Statistics''' | ||
| + | :** Volume information (WM, GM, CSF, lobes) stored in spreadsheet | ||
| + | :** Tools: ImageMath, ImageStat (UNC Slicer3 external modules) | ||
| + | :* '''5. Mesh creation''' | ||
| + | :** White matter and grey matter meshes creation | ||
| + | :** Tool: ModelMaker (Slicer3 module) | ||
| + | :* '''6. MRML scene creation for quality control''' | ||
| + | :** MRML scene creation allowing quality control for each step of the pipeline | ||
| + | :** Output images and surfaces are automatically displayed | ||
All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. | All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. | ||
| − | The user can thus compute an individual regional cortical thickness analysis by running the ' | + | The user can thus compute an individual regional cortical thickness analysis by running the 'ARCTIC' module, either within Slicer3 or as a command line. |
| + | == Download == | ||
| + | === ARCTIC download === | ||
| − | + | Source code, executables and tutorial are available on [http://www.nitrc.org/projects/arctic NITRC] | |
| − | + | === Complementary downloads === | |
| − | |||
| − | |||
| − | |||
| − | + | ==== Brain atlases ==== | |
| − | + | Four brain atlases are available on MIDAS: | |
| − | + | :* [http://www.insight-journal.org/midas/item/view/2277 Pediatric atlas] | |
| + | :* [http://www.insight-journal.org/midas/item/view/2328 Adult atlas] | ||
| + | :* [http://www.insight-journal.org/midas/item/view/2330 Elderly atlas] | ||
| + | :* [http://www.insight-journal.org/midas/item/view/2283 Primate atlas] | ||
| − | + | ==== Pediatric Brain MRI data ==== | |
| − | + | [http://insight-journal.org/midas/community/view/24 Data of 2 autistic children and 2 normal controls] (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation. | |
| − | === | + | ==== Tutorials ==== |
| + | *'''ARCTIC tutorial''' : end-to-end Slicer3 module to perform automatic regional cortical thickness analysis[[Media:ARCTIC-Slicer3-Tutorial.ppt| [ppt]]][[Media:ARCTIC-Slicer3-Tutorial.pdf| [pdf]]] | ||
| + | **1st Prize: NAMIC tutorial contest AHM 2009 | ||
| + | **2nd Prize: NAMIC tutorial contest summer project week 2009 | ||
| + | *'''UNC Modules tutorial''' : UNC Slicer3 modules to perform regional cortical thickness analysis step by step[[Media:UNC_Modules-Slicer3-Tutorial.ppt| [ppt]]][[Media:UNC_Modules-Slicer3-Tutorial.pdf| [pdf]]] | ||
| − | + | *'''[https://www.slicer.org/wiki/Modules:ARCTIC-Documentation-3.6 Online documentation within Slicer 3.6]''' | |
| − | + | <center> | |
| − | + | {| | |
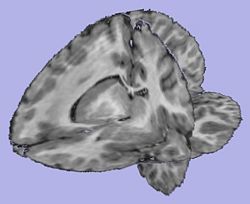
| − | + | |[[Image:T1Image.jpg|thumb|250px|T1-weighted skull-stripped image]] | |
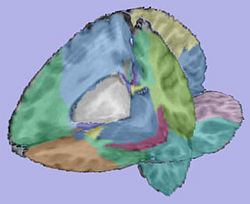
| + | |valign="center"|[[Image:Parcellation.jpg|thumb|250px|Parcellation image]] | ||
| + | |valign="center"|[[Image:WMThickness.jpg|thumb|250px|Cortical thickness on WM surface]] | ||
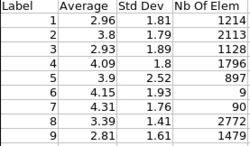
| + | |valign="center"|[[Image:ThicknessInformation.jpg|thumb|250px|Cortical thickness information]] | ||
| + | |} | ||
| + | </center> | ||
| − | + | == Pipeline validation == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Analysis on a small pediatric dataset === | === Analysis on a small pediatric dataset === | ||
| − | Tests have been computed on a small pediatric dataset | + | Tests have been computed on a small pediatric dataset which includes 2 year-old and 4 year-old cases. |
| − | :* 16 | + | :* 16 autistic cases |
:* 1 developmental delay | :* 1 developmental delay | ||
:* 3 normal control | :* 3 normal control | ||
| − | === | + | === Comparison to state of the art === |
| − | We would like to | + | We would like to compare our pipeline with FreeSurfer. We have thus performed a [[DBP2:UNC:Cortical_Thickness_Comparison | regional statistical analysis]] based on Pearson's correlation on an adult dataset (FreeSurfer's publicly available dataset) including 40 cases. We also performed [[DBP2:UNC:Cortical_Thickness_Comparison#Tests_on_a_longitudinal_pediatric_autism_study | tests on a longitudinal autism study]] of 86 subject aged 2-4 years. |
| − | + | == Planning == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Done === | === Done === | ||
:* Workflow for individual analysis (Slicer3 external module using BatchMake) | :* Workflow for individual analysis (Slicer3 external module using BatchMake) | ||
:* 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC" | :* 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC" | ||
| + | :* Pediatric and adult brain atlases available to the community via MIDAS | ||
| + | :* ARCTIC available to the community via NITRC: executables (UNC external modules for Slicer3) and Tutorial dataset | ||
| + | :** New version including quality control through MRML scene, and WM, GM models generation | ||
| + | :** Mac and windows executables also available on to the community through NITRC | ||
| + | :** Input images orientation automatically managed by ARCTIC | ||
| + | :* ARCTIC source code (SVN, CVS) available to the community | ||
| + | :* ARCTIC executable can be downloaded directly within Slicer3 using the extension wizard | ||
| + | :* Comparison to FreeSurfer (40 cases dataset): pearson correlation analysis | ||
| − | === | + | == References == |
| − | |||
| − | |||
| − | |||
| − | + | *Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet, C., & Piven, J., Early brain overgrowth in autism results from an increase in cortical surface area before age 2, Arch Gen Psychiatry. 2011;68(5):1-10 | |
| − | + | *H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009. | |
| + | *C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract | ||
Latest revision as of 17:46, 10 July 2017
Home < DBP2:UNC:Regional Cortical Thickness PipelineBack to UNC Cortical Thickness Roadmap
Contents
Objective
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness.
This page describes the related pipeline with its basic components, as well as its validation.
Pipeline overview
A Slicer3 high-level module for individual cortical thickness analysis has been developed: ARCTIC (Automatic Regional Cortical ThICkness)
Input: RAW images (T1-weighted, T2-weighted, PD-weighted images)
- 1. Tissue segmentation
- Probabilistic atlas-based automatic tissue segmentation via an Expectation-Maximization scheme
- Tool: itkEMS (UNC Slicer3 external module)
- 2. Regional atlas deformable registration
- 2.1 Skull stripping using previously computed tissue segmentation label image
- Tool: SegPostProcess (UNC Slicer3 external module)
- 2.2 T1-weighted atlas deformable registration
- B-spline pipeline registration
- Tool: RegisterImages (Slicer3 module)
- 2.3. Applying transformation to the parcellation map
- Tool: ResampleVolume2 (Slicer3 module)
- 2.1 Skull stripping using previously computed tissue segmentation label image
- 3. Cortical thickness measurement
- Sparse asymmetric local cortical thickness
- Tool: CortThick (UNC Slicer3 external module)
- 4. Statistics
- Volume information (WM, GM, CSF, lobes) stored in spreadsheet
- Tools: ImageMath, ImageStat (UNC Slicer3 external modules)
- 5. Mesh creation
- White matter and grey matter meshes creation
- Tool: ModelMaker (Slicer3 module)
- 6. MRML scene creation for quality control
- MRML scene creation allowing quality control for each step of the pipeline
- Output images and surfaces are automatically displayed
- 1. Tissue segmentation
All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. The user can thus compute an individual regional cortical thickness analysis by running the 'ARCTIC' module, either within Slicer3 or as a command line.
Download
ARCTIC download
Source code, executables and tutorial are available on NITRC
Complementary downloads
Brain atlases
Four brain atlases are available on MIDAS:
Pediatric Brain MRI data
Data of 2 autistic children and 2 normal controls (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation.
Tutorials
- ARCTIC tutorial : end-to-end Slicer3 module to perform automatic regional cortical thickness analysis [ppt] [pdf]
- 1st Prize: NAMIC tutorial contest AHM 2009
- 2nd Prize: NAMIC tutorial contest summer project week 2009
- UNC Modules tutorial : UNC Slicer3 modules to perform regional cortical thickness analysis step by step [ppt] [pdf]
Pipeline validation
Analysis on a small pediatric dataset
Tests have been computed on a small pediatric dataset which includes 2 year-old and 4 year-old cases.
- 16 autistic cases
- 1 developmental delay
- 3 normal control
Comparison to state of the art
We would like to compare our pipeline with FreeSurfer. We have thus performed a regional statistical analysis based on Pearson's correlation on an adult dataset (FreeSurfer's publicly available dataset) including 40 cases. We also performed tests on a longitudinal autism study of 86 subject aged 2-4 years.
Planning
Done
- Workflow for individual analysis (Slicer3 external module using BatchMake)
- 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC"
- Pediatric and adult brain atlases available to the community via MIDAS
- ARCTIC available to the community via NITRC: executables (UNC external modules for Slicer3) and Tutorial dataset
- New version including quality control through MRML scene, and WM, GM models generation
- Mac and windows executables also available on to the community through NITRC
- Input images orientation automatically managed by ARCTIC
- ARCTIC source code (SVN, CVS) available to the community
- ARCTIC executable can be downloaded directly within Slicer3 using the extension wizard
- Comparison to FreeSurfer (40 cases dataset): pearson correlation analysis
References
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet, C., & Piven, J., Early brain overgrowth in autism results from an increase in cortical surface area before age 2, Arch Gen Psychiatry. 2011;68(5):1-10
- H.C. Hazlett, C. Vachet, C. Mathieu, M. Styner, J. Piven, Use of the Slicer3 Toolkit to Produce Regional Cortical Thickness Measurement of Pediatric MRI Data, presented at the 8th Annual International Meeting for Autism Research (IMFAR) Chicago, IL 2009.
- C. Mathieu, C. Vachet, H.C. Hazlett, G. Geric, J. Piven, and M. Styner, ARCTIC – Automatic Regional Cortical ThICkness Tool, UNC Radiology Research Day 2009 abstract