Difference between revisions of "Collaboration:UWA-Perth"
m (Fix MediaWiki table formatting issue discovered while converting to GitHub Flavored Markdown using pandoc (via https://github.com/outofcontrol/mediawiki-to-gfm)) Tag: 2017 source edit |
|||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 12: | Line 12: | ||
Collaboration with [http://www.mech.uwa.edu.au/ISML/people.htm Karol Miller, Adam Wittek and Grand Joldes] of the University of Western Australia. | Collaboration with [http://www.mech.uwa.edu.au/ISML/people.htm Karol Miller, Adam Wittek and Grand Joldes] of the University of Western Australia. | ||
| − | |||
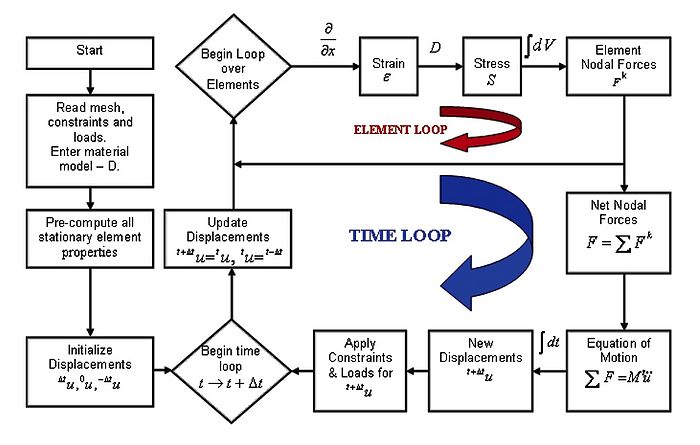
[[Image:TLED-grabJPG.jpg|700px]] | [[Image:TLED-grabJPG.jpg|700px]] | ||
| − | + | ||
| − | |||
Flow chart of the finite element algorithm with Total Lagrange Explicit Dynamics (TLED) for computing soft organ deformation developed at ISML. Detailed description is given in the reference [5]. | Flow chart of the finite element algorithm with Total Lagrange Explicit Dynamics (TLED) for computing soft organ deformation developed at ISML. Detailed description is given in the reference [5]. | ||
| − | |||
==Goals of the Project== | ==Goals of the Project== | ||
Mathematical modelling and computer simulation have proved tremendously successful in engineering. One of the greatest challenges for mechanists is to extend the success of computational mechanics to fields outside traditional engineering, in particular to biology, biomedical sciences, and medicine. For example, continuum mechanics models provide a rational basis for analysing biomedical images by constraining the solution to biologically reasonable motions and processes. Biomechanical modelling can also provide clinically important information about the physical status of the underlying biology, integrating information across molecular, tissue, organ, and organism scales. | Mathematical modelling and computer simulation have proved tremendously successful in engineering. One of the greatest challenges for mechanists is to extend the success of computational mechanics to fields outside traditional engineering, in particular to biology, biomedical sciences, and medicine. For example, continuum mechanics models provide a rational basis for analysing biomedical images by constraining the solution to biologically reasonable motions and processes. Biomechanical modelling can also provide clinically important information about the physical status of the underlying biology, integrating information across molecular, tissue, organ, and organism scales. | ||
| − | We intend to contribute to Na-MIC by providing algorithms for computing the intra-operative brain deformations for image-guided neurosurgery. We treat the brain shift as a continuum mechanics problem involving finite deformations and solve it using non-linear finite element procedures. We use the finite element procedures (Total Lagrangian formulation and Dynamic Relaxation that combines explicit integration in time domain with mass proportional damping) that do not require iterations even when applied to non-linear problems and, therefore, make it possible to compute the intra-operative brain deformations in real time: under 40 s on a standard PC and under 4 s on a graphics processing unit GPU (we used NVIDIA Tesla C870) [https://commerce.metapress.com/content/u054680542xw31v7/resource-secured/?target=fulltext.pdf&sid=3hfkdh45m5b54d55lwnqmhzt&sh=www.springerlink.com] [http://www.mech.uwa.edu.au/ISML/reports/isml_no_03_2009.pdf], [http://www.mech.uwa.edu.au/ISML/reports/isml-no-01-2009.pdf], [5], [ | + | We intend to contribute to Na-MIC by providing algorithms for computing the intra-operative brain deformations for image-guided neurosurgery. We treat the brain shift as a continuum mechanics problem involving finite deformations and solve it using non-linear finite element procedures. We use the finite element procedures (Total Lagrangian formulation and Dynamic Relaxation that combines explicit integration in time domain with mass proportional damping) that do not require iterations even when applied to non-linear problems and, therefore, make it possible to compute the intra-operative brain deformations in real time: under 40 s on a standard PC and under 4 s on a graphics processing unit GPU (we used NVIDIA Tesla C870) [https://commerce.metapress.com/content/u054680542xw31v7/resource-secured/?target=fulltext.pdf&sid=3hfkdh45m5b54d55lwnqmhzt&sh=www.springerlink.com] [http://www.mech.uwa.edu.au/ISML/reports/isml_no_03_2009.pdf], [http://www.mech.uwa.edu.au/ISML/reports/isml-no-01-2009.pdf], [5], [6], [9], [17-20]. |
==Current Progress== | ==Current Progress== | ||
| − | The algorithms have been already implemented in C/C++ using Visual Studio and MFC. The GPU implementation was done using NVIDIA's Compute Unified Device Architecture (CUDA) [ | + | The algorithms have been already implemented in C/C++ using Visual Studio and MFC. The GPU implementation was done using NVIDIA's Compute Unified Device Architecture (CUDA) [2]. Matlab is used for visualizing the results. |
| − | The algorithms' accuracy in predicting the intraoperative brain deformation has been successfully verified against the intraoperative MRIs acquired during the craniotomy [2] | + | The algorithms' accuracy in predicting the intraoperative brain deformation has been successfully verified against the intraoperative MRIs acquired during the craniotomy [1] and [2]. |
| + | |||
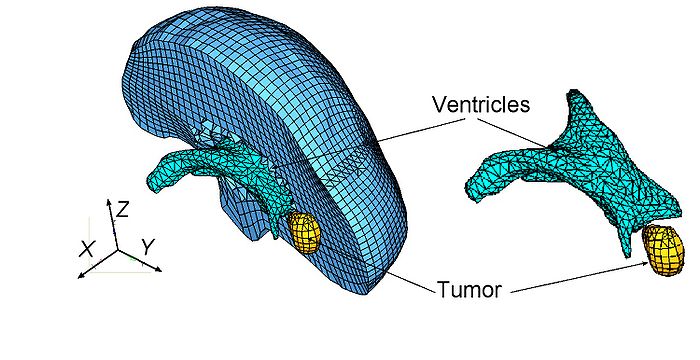
| + | [[Image:ModelView-JPEG.jpg|700px]] | ||
| + | |||
| + | Typical example of a patient-specific finite element mesh applied in study [1] to predict the deformation field within the brain due to craniotomy-induced brain shift. For this mesh, our specialised non-linear finite element algorithms for soft tissue modelling made it possible to compute the deformation field in 30 s using a PC (Intel E6850 dual core 3.00 GHz processor, 4 GB of internal memory, Windows XP operating system) and 4 s using a GPU (NVIDIA Tesla C870). | ||
==Publications== | ==Publications== | ||
| Line 35: | Line 36: | ||
#Joldes, G. R., Wittek, A. and Miller, K. (2009a) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6V29-4WMDHFK-3&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1159070912&_rerunOrigin=google&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cc5a7bf863d9aba9d45b165e173b1591 Computation of intra-operative brain shift using dynamic relaxation]. Computer Methods in Applied Mechanics and Engineering 198, 3313-3320. | #Joldes, G. R., Wittek, A. and Miller, K. (2009a) [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6V29-4WMDHFK-3&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1159070912&_rerunOrigin=google&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cc5a7bf863d9aba9d45b165e173b1591 Computation of intra-operative brain shift using dynamic relaxation]. Computer Methods in Applied Mechanics and Engineering 198, 3313-3320. | ||
#Miller, K., Wittek, A., Joldes, G., Horton, A., Dutta Roy, T., Berger, J. and Morriss, L. (2009) [http://www3.interscience.wiley.com/journal/122380937/abstract?CRETRY=1&SRETRY=0 Modelling brain deformations for computer-integrated neurosurgery (invited review)]. Communications in Numerical Methods in Engineering 26, 111-138. | #Miller, K., Wittek, A., Joldes, G., Horton, A., Dutta Roy, T., Berger, J. and Morriss, L. (2009) [http://www3.interscience.wiley.com/journal/122380937/abstract?CRETRY=1&SRETRY=0 Modelling brain deformations for computer-integrated neurosurgery (invited review)]. Communications in Numerical Methods in Engineering 26, 111-138. | ||
| − | #Joldes, G., Wittek, A. and Miller, K. [http://www.radiologysource.org/periodicals/medima/article/S1361-8415(08)00142-4/abstract Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation.] Med Image Anal. 2009 Dec;13(6):912- | + | #Joldes, G., Wittek, A. and Miller, K. [http://www.radiologysource.org/periodicals/medima/article/S1361-8415(08)00142-4/abstract Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation.] Med Image Anal. 2009 Dec;13(6):912-919. PubMed PMID: 19152791; PubMed Central PMCID: PMC2783832. |
| − | #Wittek A, Hawkins T, Miller K. [http://www.na-mic.org/publications/item/view/1554 On the | + | #Wittek A., Hawkins T., Miller K. [http://www.na-mic.org/publications/item/view/1554 On the Unimportance of Constitutive Models in Computing Brain Deformation for Image-guided Surgery.] Biomech Model Mechanobiol. 2009 Feb;8(1):77-84. PMID: 18246376. PMCID: PMC3224703. |
#Joldes, G., Wittek, A. and Miller, K. [[http://www3.interscience.wiley.com/journal/114293156/abstract?CRETRY=1&SRETRY=0 An efficient hourglass control implementation for the uniform strain hexahedron using the Total Lagrangian formulation.] Communications in Numerical Methods in Engineering, 2008;24:1315-1323. | #Joldes, G., Wittek, A. and Miller, K. [[http://www3.interscience.wiley.com/journal/114293156/abstract?CRETRY=1&SRETRY=0 An efficient hourglass control implementation for the uniform strain hexahedron using the Total Lagrangian formulation.] Communications in Numerical Methods in Engineering, 2008;24:1315-1323. | ||
#Miller, K., Joldes, G., Lance, D., Wittek, A. [http://www3.interscience.wiley.com/journal/112692131/abstract Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation.] Communications in Numerical Methods in Engineering. 2007;23:121-134. | #Miller, K., Joldes, G., Lance, D., Wittek, A. [http://www3.interscience.wiley.com/journal/112692131/abstract Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation.] Communications in Numerical Methods in Engineering. 2007;23:121-134. | ||
| − | #Wittek A, Miller K, Kikinis R, Warfield S. [http://www.na-mic.org/publications/item/view/94 Patient-specific | + | #Wittek A., Miller K., Kikinis R., Warfield S.K. [http://www.na-mic.org/publications/item/view/94 Patient-specific Model of Brain Deformation: Application to Medical Image Registration..] J Biomech. 2007;40(4):919-29. PMID: 16678834. |
#Joldes, G. R., Wittek, A., Miller, K. (2007) Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation, in Proceedings of Computational Biomechanics for Medicine Workshop, workshop associated with International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2007, Brisbane, Australia, ISBN 13: 978 0 643 09517 5, pp. 65-73. | #Joldes, G. R., Wittek, A., Miller, K. (2007) Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation, in Proceedings of Computational Biomechanics for Medicine Workshop, workshop associated with International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2007, Brisbane, Australia, ISBN 13: 978 0 643 09517 5, pp. 65-73. | ||
#Hawkins, T., Wittek, A. and Miller, K. (2006) Comparison of constitutive models of brain tissue for non-rigid image registration, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages. | #Hawkins, T., Wittek, A. and Miller, K. (2006) Comparison of constitutive models of brain tissue for non-rigid image registration, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages. | ||
| Line 49: | Line 50: | ||
#Miller, K. Hawkins, T. and Wittek, A. (2006) Linear versus non-linear computation of the brain shift, in CD Proceedings of the 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 888-893. | #Miller, K. Hawkins, T. and Wittek, A. (2006) Linear versus non-linear computation of the brain shift, in CD Proceedings of the 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 888-893. | ||
#Miller, K. and Wittek, A. (2006) [[http://www.mech.uwa.edu.au/ISML/CBM_06_miller.pdf Neuroimage registration as displacement - zero traction problem of solid mechanics.] Lead Lecture in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 1-12. | #Miller, K. and Wittek, A. (2006) [[http://www.mech.uwa.edu.au/ISML/CBM_06_miller.pdf Neuroimage registration as displacement - zero traction problem of solid mechanics.] Lead Lecture in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 1-12. | ||
| − | #Wittek A, Kikinis R, Warfield S, Miller K. [http://www.na-mic.org/publications/item/view/216 Brain | + | #Wittek A, Kikinis R, Warfield S, Miller K. [http://www.na-mic.org/publications/item/view/216 Brain Shift Computation using a Fully Nonlinear Biomechanical Model.] Int Conf Med Image Comput Comput Assist Interv. 2005;8(Pt 2):583-90. PMID: 16686007. |
===In Press=== | ===In Press=== | ||
Latest revision as of 07:06, 11 April 2023
Home < Collaboration:UWA-PerthBack to NA-MIC External Collaborations
Real Time Computer Simulation of Human Soft Organ Deformation for Computer Assisted Surgery
Key Personnel
- UWA: Karol Miller, PI
- NA-MIC: Ron Kikinis
The proposed research will develop computational framework, which will allow calculation of soft organ (brain, liver, kidney, prostate, etc.) deformation during surgical operations in real time. Fully non-linear material models and geometrically nonlinear finite element formulation will be used. The fundamental technology developed within this project: physically (or mechanically) realistic modeling and real time computer simulation of soft organ deformation, will have applications in many areas of computer assisted surgery, such as intra-operative, real time non-rigid registration and virtual reality surgeon training and operation planning systems with force and tactile feedback.
Click here to learn more about ISML.
Collaboration with Karol Miller, Adam Wittek and Grand Joldes of the University of Western Australia.
Flow chart of the finite element algorithm with Total Lagrange Explicit Dynamics (TLED) for computing soft organ deformation developed at ISML. Detailed description is given in the reference [5].
Goals of the Project
Mathematical modelling and computer simulation have proved tremendously successful in engineering. One of the greatest challenges for mechanists is to extend the success of computational mechanics to fields outside traditional engineering, in particular to biology, biomedical sciences, and medicine. For example, continuum mechanics models provide a rational basis for analysing biomedical images by constraining the solution to biologically reasonable motions and processes. Biomechanical modelling can also provide clinically important information about the physical status of the underlying biology, integrating information across molecular, tissue, organ, and organism scales.
We intend to contribute to Na-MIC by providing algorithms for computing the intra-operative brain deformations for image-guided neurosurgery. We treat the brain shift as a continuum mechanics problem involving finite deformations and solve it using non-linear finite element procedures. We use the finite element procedures (Total Lagrangian formulation and Dynamic Relaxation that combines explicit integration in time domain with mass proportional damping) that do not require iterations even when applied to non-linear problems and, therefore, make it possible to compute the intra-operative brain deformations in real time: under 40 s on a standard PC and under 4 s on a graphics processing unit GPU (we used NVIDIA Tesla C870) [1] [2], [3], [5], [6], [9], [17-20].
Current Progress
The algorithms have been already implemented in C/C++ using Visual Studio and MFC. The GPU implementation was done using NVIDIA's Compute Unified Device Architecture (CUDA) [2]. Matlab is used for visualizing the results. The algorithms' accuracy in predicting the intraoperative brain deformation has been successfully verified against the intraoperative MRIs acquired during the craniotomy [1] and [2].
Typical example of a patient-specific finite element mesh applied in study [1] to predict the deformation field within the brain due to craniotomy-induced brain shift. For this mesh, our specialised non-linear finite element algorithms for soft tissue modelling made it possible to compute the deformation field in 30 s using a PC (Intel E6850 dual core 3.00 GHz processor, 4 GB of internal memory, Windows XP operating system) and 4 s using a GPU (NVIDIA Tesla C870).
Publications
For complete list of our publication visit here.
- Joldes, G. R., Wittek, A., Couton, M., Warfield, S. K. and Miller, K. (2009) Real-time prediction of brain shift using nonlinear finite element algorithms In Proc. 12 International Conference on Medical Image Computing and Computer Assisted Intervention MICCAI 2009, Lecture Notes in Computer Science 5761, 300-307.
- Joldes, G., Wittek, A., Miller, K. (2009) Real-Time Nonlinear Finite Element Computations on GPU for Surgical Simulation. Intelligent Systems for Medicine Laboratory, University of Western Australia, Report # ISML/03/2009, 21 pages.
- Wittek, A. , Joldes, G., Miller, K. (2009) Evaluation of the Accuracy of Nonlinear Finite Element Modelling for Predicting Intraoperative Brain Deformations: Study of Six Craniotomy Cases. Intelligent Systems for Medicine Laboratory, University of Western Australia, Report # ISML/01/2009, 18 pages.
- Joldes, G. R., Wittek, A. and Miller, K. (2009a) Computation of intra-operative brain shift using dynamic relaxation. Computer Methods in Applied Mechanics and Engineering 198, 3313-3320.
- Miller, K., Wittek, A., Joldes, G., Horton, A., Dutta Roy, T., Berger, J. and Morriss, L. (2009) Modelling brain deformations for computer-integrated neurosurgery (invited review). Communications in Numerical Methods in Engineering 26, 111-138.
- Joldes, G., Wittek, A. and Miller, K. Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation. Med Image Anal. 2009 Dec;13(6):912-919. PubMed PMID: 19152791; PubMed Central PMCID: PMC2783832.
- Wittek A., Hawkins T., Miller K. On the Unimportance of Constitutive Models in Computing Brain Deformation for Image-guided Surgery. Biomech Model Mechanobiol. 2009 Feb;8(1):77-84. PMID: 18246376. PMCID: PMC3224703.
- Joldes, G., Wittek, A. and Miller, K. [An efficient hourglass control implementation for the uniform strain hexahedron using the Total Lagrangian formulation. Communications in Numerical Methods in Engineering, 2008;24:1315-1323.
- Miller, K., Joldes, G., Lance, D., Wittek, A. Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation. Communications in Numerical Methods in Engineering. 2007;23:121-134.
- Wittek A., Miller K., Kikinis R., Warfield S.K. Patient-specific Model of Brain Deformation: Application to Medical Image Registration.. J Biomech. 2007;40(4):919-29. PMID: 16678834.
- Joldes, G. R., Wittek, A., Miller, K. (2007) Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation, in Proceedings of Computational Biomechanics for Medicine Workshop, workshop associated with International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2007, Brisbane, Australia, ISBN 13: 978 0 643 09517 5, pp. 65-73.
- Hawkins, T., Wittek, A. and Miller, K. (2006) Comparison of constitutive models of brain tissue for non-rigid image registration, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages.
- Horton, A., Wittek, A. and K. Miller (2006) Computer simulation of brain shift using an element free Galerkin method, in CD Proceedings of 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp.906-911.
- Horton, A., Wittek, A. and K. Miller Towards meshless methods for surgery simulation. Linear versus non-linear computation of the brain shift. Proceedings of Computational Biomechanics for Medicine Workshop, workshop associated with International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 32-40.
- Joldes, G., Wittek, A. and Miller, K. (2006) Improved linear tetrahedral element for surgery simulation. Proceedings of Computational Biomechanics for Medicine Workshop, workshop associated with International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 52-63.
- Joldes, G., Wittek, A. and Miller, K. (2006) Towards non-linear finite element, computations in real time, in CD Proceedings of 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 894-899.
- Miller, K., Joldes, G. and Wittek, A. (2006) New finite element algorithm for surgical simulation, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages
- Miller, K. Hawkins, T. and Wittek, A. (2006) Linear versus non-linear computation of the brain shift, in CD Proceedings of the 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 888-893.
- Miller, K. and Wittek, A. (2006) [Neuroimage registration as displacement - zero traction problem of solid mechanics. Lead Lecture in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 1-12.
- Wittek A, Kikinis R, Warfield S, Miller K. Brain Shift Computation using a Fully Nonlinear Biomechanical Model. Int Conf Med Image Comput Comput Assist Interv. 2005;8(Pt 2):583-90. PMID: 16686007.