Difference between revisions of "EMSegmenter"
| (12 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
Tracing the history of the EMSegmenter... | Tracing the history of the EMSegmenter... | ||
| − | ===1995=== | + | ===1993-1995=== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | === | + | ====Development==== |
| − | + | The EM segmenter grew out of a collaboration between Sandy Wells, Ron Kikinis and Martha Shenton in about 1993. The goal was to get good automatic segmentations of white matter and gray matter from T1 weighted MRI. The biggest difficulty was the intensity inhomogeneities, or "shading", artifact in the images that was due to the MRI scanner used for research at that time. The effect of the artifact was that a single threshold could not be used to separate white matter and gray matter. | |
| − | |||
| − | |||
| − | |||
| + | Various approaches to the problem were tried, some giving good results, but there were remaining imperfections in the results. | ||
| + | Eventually, we decided to construct an explicit representation of the intensity artifact, and attempt to recover the artifact and the segmentation simultaneously. | ||
| + | We chose the Expectation Maximization (EM) algorithm, a statistical estimation method that is used when some data is considered to be | ||
| + | "missing". The result was an iterative algorithm that alternates between two steps. | ||
| − | + | In the "E" step, the probability of the tissue label at each voxel is estimated, given the image data and the current estimate of the | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | In the "E" step, the probability of the tissue label at each voxel is | ||
| − | estimated, given the image data and the current estimate of the | ||
intensity artifact. | intensity artifact. | ||
| − | In the "M" step, the intensity artifact is re-estimated, given the | + | In the "M" step, the intensity artifact is re-estimated, given the image data and current estimate of the tissue label probabilities. |
| − | image data and current estimate of the tissue label probabilities. | ||
| − | The EM segmenter proved to be very robust to shading artifacts, | + | The EM segmenter proved to be very robust to shading artifacts, but in addition, it was also robust to "inter-scan |
| − | but in addition, it was also robust to "inter-scan | + | inhomogeneities". With previous classification approaches to segmentation, "training" was needed on a per-scan basis, because |
| − | inhomogeneities". With previous classification approaches to | ||
| − | segmentation, "training" was needed on a per-scan basis, because | ||
of intensity changes from scan to scan. | of intensity changes from scan to scan. | ||
| − | The EM segmener was the first algorithm that could produce high | + | The EM segmener was the first algorithm that could produce high quality segmentations of white matter and gray matter from MRI, |
| − | quality segmentations of white matter and gray matter from MRI, | + | with no manual intervention needed on a per case basis. This proved to be very valuable in a large longitudinal study of MS in the |
| − | with no manual intervention needed on a per case basis. This proved | ||
| − | to be very valuable in a large longitudinal study of MS in the | ||
period 1994 - 1995. | period 1994 - 1995. | ||
| + | {| | ||
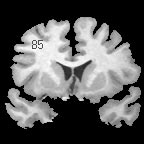
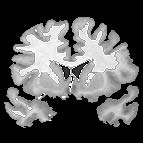
| + | |+ '''Fig 1. EM Segmentation: Cross-Sections of Segmentations (1993)''' | ||
| + | |valign="top"|[[Image:one.jpg|thumb|252px|slice of T1 weighted mr (right temporal lobe has bad "shading")]] | ||
| + | |valign="top"|[[Image:two.jpg|thumb|252px|threshoding result]] | ||
| + | |valign="top"|[[Image:three.jpg|thumb|252px|EM result]] | ||
| + | |} | ||
| − | + | {| | |
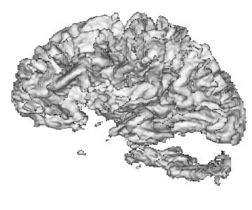
| − | + | |+ '''Fig 2. EM Segmentation: 3D Rendered Segmentations (1993)''' | |
| − | + | |valign="top"|[[Image:four.jpg|thumb|252px|3D view of segmented white matter surface from thresholding]] | |
| − | + | |valign="top"|[[Image:five.jpg|thumb|252px|3D view of segmented white matter surface from EM]] | |
| − | + | |} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {| | |
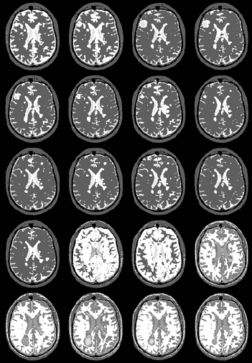
| + | |+ '''Fig 3. MS Longitudinal Study (1995)''' | ||
| + | |valign="top"|[[Image:seven.jpg|thumb|252px|longituidinal MS, one subect, segmentaiton result without EM]] | ||
| + | |valign="top"|[[Image:eight.jpg|thumb|252px|Results with EM]] | ||
| + | |} | ||
| − | + | {| | |
| − | [[Image: | + | |+ '''Fig 4. Surface Coil EM Segmentations(1995)''' |
| + | |valign="top"|[[Image:nine.jpg|thumb|252px|surface coil image]] | ||
| + | |valign="top"|[[Image:ten.jpg|thumb|252px|surface coil image corrected by EM algorithm]] | ||
| + | |} | ||
| − | + | ====Publication==== | |
| − | + | Adaptive Segmentation of MRI Data. WM Wells III, WEL Grimson, R Kikinis, FA Jolesz. IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 15, NO. 4, AUGUST 1996 | |
| − | + | ===1995-1998=== | |
| − | |||
| − | + | ====Development==== | |
| + | From 1995-1998, MRF models were incorporated into the EM segmenter via a Mean Field Solver. The resultant segmentations more robust to noise. | ||
| − | + | ====Publication==== | |
| + | Enhanced Spatial Priors for Segmentation of Magnetic Resonance Imagery. T. Kapur, W.E.L. Grimson, W. M. Wells III, R. Kikinis, MICCAI, Cambridge, MA, Octobery 1998. | ||
| − | + | ===2000-2007 === | |
| + | From 2000 to now, spatial priors were added to the EM segmenter. Specifically, | ||
| + | * Added the use of anatomical atlases of specific brain parts, e.g., the hippocampus : started to be a brain "parcellator" | ||
| + | * Added simultaneous registration of atlas and subject | ||
| + | * developed a hierarchial method for parcellation and validated it on schizophrenia data | ||
| + | * developed a mean-field level-set post-processor that is effective for reducing the effects of noise. | ||
| − | |||
| − | + | ===Additional Slides=== | |
| + | (From Kilian) | ||
| − | + | * [[IMAGE:Ron-ISBI-07.zip| Here]] are the research related slides | |
| − | + | * The [[Media:EMFeedback3.ppt | Feedback slides]] featuring the transition from Slicer 2 to Slicer 3 were generated by Brad and Kilian. | |
Latest revision as of 16:41, 14 April 2007
Home < EMSegmenterContents
History
Tracing the history of the EMSegmenter...
1993-1995
Development
The EM segmenter grew out of a collaboration between Sandy Wells, Ron Kikinis and Martha Shenton in about 1993. The goal was to get good automatic segmentations of white matter and gray matter from T1 weighted MRI. The biggest difficulty was the intensity inhomogeneities, or "shading", artifact in the images that was due to the MRI scanner used for research at that time. The effect of the artifact was that a single threshold could not be used to separate white matter and gray matter.
Various approaches to the problem were tried, some giving good results, but there were remaining imperfections in the results. Eventually, we decided to construct an explicit representation of the intensity artifact, and attempt to recover the artifact and the segmentation simultaneously.
We chose the Expectation Maximization (EM) algorithm, a statistical estimation method that is used when some data is considered to be "missing". The result was an iterative algorithm that alternates between two steps.
In the "E" step, the probability of the tissue label at each voxel is estimated, given the image data and the current estimate of the intensity artifact.
In the "M" step, the intensity artifact is re-estimated, given the image data and current estimate of the tissue label probabilities.
The EM segmenter proved to be very robust to shading artifacts, but in addition, it was also robust to "inter-scan inhomogeneities". With previous classification approaches to segmentation, "training" was needed on a per-scan basis, because of intensity changes from scan to scan.
The EM segmener was the first algorithm that could produce high quality segmentations of white matter and gray matter from MRI, with no manual intervention needed on a per case basis. This proved to be very valuable in a large longitudinal study of MS in the period 1994 - 1995.
Publication
Adaptive Segmentation of MRI Data. WM Wells III, WEL Grimson, R Kikinis, FA Jolesz. IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 15, NO. 4, AUGUST 1996
1995-1998
Development
From 1995-1998, MRF models were incorporated into the EM segmenter via a Mean Field Solver. The resultant segmentations more robust to noise.
Publication
Enhanced Spatial Priors for Segmentation of Magnetic Resonance Imagery. T. Kapur, W.E.L. Grimson, W. M. Wells III, R. Kikinis, MICCAI, Cambridge, MA, Octobery 1998.
2000-2007
From 2000 to now, spatial priors were added to the EM segmenter. Specifically,
- Added the use of anatomical atlases of specific brain parts, e.g., the hippocampus : started to be a brain "parcellator"
- Added simultaneous registration of atlas and subject
- developed a hierarchial method for parcellation and validated it on schizophrenia data
- developed a mean-field level-set post-processor that is effective for reducing the effects of noise.
Additional Slides
(From Kilian)
- File:Ron-ISBI-07.zip are the research related slides
- The Feedback slides featuring the transition from Slicer 2 to Slicer 3 were generated by Brad and Kilian.