Difference between revisions of "Slicer3.2:Training"
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) |
||
| Line 62: | Line 62: | ||
| style="background:#9BF2C5; color:black" align="Center"| '''1.6''' | | style="background:#9BF2C5; color:black" align="Center"| '''1.6''' | ||
| style="background:#D1FFF9; color:black"| [http://www.nitrc.org/frs/download.php/513/Slicer3Training_WhiteMatterLesions.ppt.pdf Detecting white matter lesions in lupus] <br> | | style="background:#D1FFF9; color:black"| [http://www.nitrc.org/frs/download.php/513/Slicer3Training_WhiteMatterLesions.ppt.pdf Detecting white matter lesions in lupus] <br> | ||
| − | This tutorial consists of five MR scans of patients with lupus. | + | This tutorial consists of five MR scans of patients with lupus. |
| style="background:#D1FFF9; color:black"|[http://www.nitrc.org/frs/download.php/512/20081114_public_lupus_data_tutorial_release.tgz Lupus-Tutorial-Data ] | | style="background:#D1FFF9; color:black"|[http://www.nitrc.org/frs/download.php/512/20081114_public_lupus_data_tutorial_release.tgz Lupus-Tutorial-Data ] | ||
| style="background:#C3D1C3; color:black" align="Center"| [[Image:Lupus2.png|200px|Lesion Segmentation]] | | style="background:#C3D1C3; color:black" align="Center"| [[Image:Lupus2.png|200px|Lesion Segmentation]] | ||
Revision as of 16:31, 16 November 2008
Home < Slicer3.2:TrainingWelcome to the 3D Slicer3.2 User Training 101
This page is under construction The information on this page applies to 3D Slicer version 3. If you are looking for materials about Slicer version 2, please go to the following location.
|
Slicer Logo | |
Software Installation
The Slicer download page contains links for downloading the different versions of Slicer 3.
Training Compendium
| Category/# | Tutorial | Sample Data | Image |
| 1.1 | Data Loading and Visualization in Slicer3 | SlicerSampleVisualization.tar.gz SlicerSampleVisualization.zip |
|
| 1.2 | EM Segmentation Course |
AutomaticSegmentation.tar.gz Older Materials: 1 |

|
| 1.3 | Affine and deformable registration in Slicer 3 | SlicerSampleRegistration.tgz | 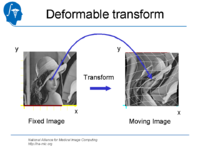
|
| 1.4 | Processing of DWI and DTI data in Slicer3 (pdf) (under construction) Processing of DWI and DTI data in Slicer3 (ppt) |
Download diffusion MRI data for the tutorial (Slicer3-diffusion-Tutorial material.zip) |
|
| 1.5 | Detecting subtle change in pathology This tutorial consists of two MR scans of a patient with meningioma. |
ChageTracker-Tutorial-Data.xcat | 
|
| 1.6 | Detecting white matter lesions in lupus This tutorial consists of five MR scans of patients with lupus. |
Lupus-Tutorial-Data | 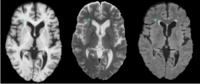
|
| 2.1 | Plug-ins for Slicer3: Course for Developers
This tutorial is intended for engineers and scientists who want to integrate external programs with Slicer3. |
HelloWorld.zip | 
|
| 2.2 | Image Guided Therapy Planning Tutorial See here for background info.
This tutorial takes the trainee through a complete workup for neurosurgical patient specific mapping |
NeurosurgicalPlanningTutorialData.zip | 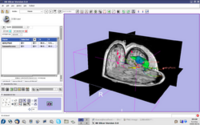
|
| 3.1 | Slicer3 as a research tool for image guided therapy research (IGT)
This tutorial is intended for engineers and scientists who want to use Slicer 3 for IGT research. |

| |
| 3.2 | SPL-PNL Brain Atlas
This three-dimensional brain atlas dataset, derived from a volumetric, whole-head MRI scan, contains the original MRI-scan, a complete set of label maps, three-dimensional reconstructions (200+ structures) and a tutorial. See here for more information. |
SPL-PNL Brain Atlas | 
|
| 3.3 | SPL Abdominal Atlas
This three-dimensional abdominal atlas was derived from a computed tomography (CT) scan, using semi-automated image segmentation and three-dimensional reconstruction techniques. The dataset contains the original CT scan, detailed label maps, three-dimensional reconstructions and a tutorial. See here for more information. |
SPL Abdominal Atlas | 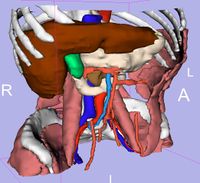
|
| 3.4 | Slicer 3 as a Research Tool for Microscopy Data
This tutorial is intended for researchers who wish to utilize Slicer 3 with microscopy data. If you are interested in more microscopy data, you can visit the Cell Centered Database for freely available data sets. |
Microscopy Sample Data (90 MB) | 
|
| 3.5 | Using XNAT Desktop and Slicer3 for remote data handling
This tutorial is intended for researchers who wish to upload local datasets to a remote XNAT repository like XNAT Central and retrieve catalog files that can be opened inside Slicer. |
|
- Category 1 = Basic functionality
- Category 2 = Advanced functionality
- Category 3 = Specialized application packages
Additional Materials
For a variety of data sets for downloading, check the following link.
Back to Training:Main
