Projects:BrainManifold
Back to Utah Algorithms
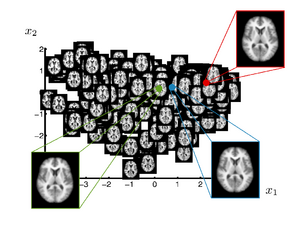
Brain Manifold Learning
This work investigates the use of manifold learning approaches in the context of brain population analysis. The goal is to construct a manifold model from a set of brain images that captures variability in shape, a parametrization of the shape space. Such a manifold model is interesting in several ways
- The low dimensional parametrization simplifies statistical analysis of populations.
- Applications to searching and browsing large database
- The manifold represents a localized Atlas. Alternative to template based applications, for example as a segmentation prior.
- Aid in clinical diagnosis. Different regions on the manifold can indicate different pathologies.
Description
In many neuroimage applications a summary or representation of a population of brain images is needed. A common approach is to build a template, or atlas, that represents a population. Recent work introduced clustering based approaches, which in a data driven fashion, compute multiple templates Each template represents a part of the population. In a different direction, researcher proposed kernel-based regression of brain images with respect to an underlying parameter. This yields a continuous curve in the space of brain images that estimates the conditional expectation of a brain image given the parameter. A natural question that arises based on these investigations is can the space spanned by a set of brain images be approximated by a low-dimensional manifold? In other words, how effectively can a low-dimensional, nonlinear model represent the variability in brain anatomy.
Key Investigators
- Utah: Samuel Gerber, Tolga Tasdizen, Sarang Joshi, Tom Fletcher, Ross Whitaker
Publications
In Print Published in MICCAI and ICCV
In Press
- S Gerber, T Tasdizen, R Whitaker, Dimensionality Reduction and Principal Surfaces via Kernel Map, ICCV 2009
- S Gerber, T Tasdizen, S Joshi, R Whitaker, On the Manifold Structure of the Space of Brain Images, MICCAI 2009