DBP2:UNC:Regional Cortical Thickness Pipeline
Back to UNC Cortical Thickness Roadmap
Contents
Objective
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness.
This page describes the related pipeline with its basic components, as well as its validation.
Pipeline overview
A Slicer3 high-level module for individual cortical thickness analysis has been developed: ARCTIC (Automatic Regional Cortical ThICkness)
Input: RAW images (T1-weighted, T2-weighted, PD-weighted images)
- 1. Tissue segmentation
- Probabilistic atlas-based automatic tissue segmentation via an Expectation-Maximization scheme
- Tool: itkEMS (UNC Slicer3 external module)
- 2. Regional atlas deformable registration
- 2.1 Skull stripping using previously computed tissue segmentation label image
- Tool: SegPostProcess (UNC Slicer3 external module)
- 2.2 T1-weighted atlas deformable registration
- B-spline pipeline registration
- Tool: RegisterImages (Slicer3 module)
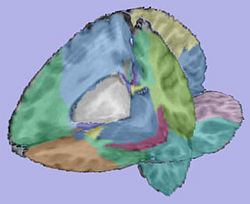
- 2.3. Applying transformation to the parcellation map
- Tool: ResampleVolume2 (Slicer3 module)
- 2.1 Skull stripping using previously computed tissue segmentation label image
- 3. Cortical Thickness
- Sparse asymmetric local cortical thickness
- Tool: CortThick (UNC Slicer3 external module)
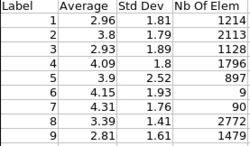
- 4. Statistics
- Volume information (WM, GM, CSF, lobes) stored in spreadsheet
- Tools: ImageMath, ImageStat (UNC Slicer3 external modules)
- 5. Mesh creation
- White matter and grey matter meshes creation
- Tool: ModelMaker (Slicer3 module)
- 6. MRML scene creation for quality control
- MRML scene creation allowing quality control for each step of the pipeline
- Output images and surfaces are automatically displayed
- 1. Tissue segmentation
All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. The user can thus compute an individual regional cortical thickness analysis by running the 'ARCTIC' module, either within Slicer3 or as a command line.
ARCTIC Download
CVS access, Executables and tutorial dataset
Available on NITRC : http://www.nitrc.org/projects/arctic/
Complementary Downloads
Brain atlases
Two brain atlases are available on MIDAS:
- Pediatric atlas: http://www.insight-journal.org/midas/item/view/2277
- Adult atlas: http://www.insight-journal.org/midas/item/view/2328
Tutorials
• ARCTIC tutorial : end-to-end Slicer3 module to perform automatic regional cortical thickness analysis [ppt] [pdf]
• UNC Modules tutorial : UNC Slicer3 modules to perform regional cortical thickness analysis step by step [ppt] [pdf]
Pipeline validation
Analysis on a small pediatric dataset
Tests have been computed on a small pediatric dataset which includes 2 year-old and 4 year-old cases.
- 16 autistic cases
- 1 developmental delay
- 3 normal control
Comparison to state of the art
We would like to compare our pipeline with FreeSurfer. We have thus started to perform a regional statistical analysis using Pearson's correlation coefficient on a pediatric dataset including 90 cases.
Two distinct groups are considered: 2 year-old cases and 4 year-old cases.
Planning
Done
- Workflow for individual analysis (Slicer3 external module using BatchMake)
- 2 Tutorials with application example on a small dataset : "How to use the UNC modules to compute the regional cortical thickness" and "How to use ARCTIC"
- Pediatric atlases available to the community through MIDAS
- ARCTIC available to the community through NITRC: executables (UNC external modules for Slicer3) and Tutorial dataset
- New version including quality control through MRML scene, and WM, GM models generation
- ARCTIC source code (SVN, CVS) available to the community
In progress
- Comparison to FreeSurfer (180 cases dataset): pearson correlation analysis
- Mac and windows executables available on to the community through NITRC
- Input images orientation problem automatically managed by ARCTIC