Difference between revisions of "2008 Winter Project Week:Fluid Mechanics Tractography"
| (4 intermediate revisions by the same user not shown) | |||
| Line 16: | Line 16: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
Computational fluid dynamics is a rich field and its application to the analysis of diffusion tensor imaging (DTI) datasets has yielded possible applications to tractography, image registration, and white matter pathology. We are developing several useful and novel diffusion tensor imaging (DTI) analysis algorithms modeled on the principles of fluid mechanics for inclusion within the NA-MIC framework. The goal of this project is to develop these methods, make them compatible with the NA-MIC ITK-based software infrastructure (i.e. Slicer), and promote their dissemination to the scientific community. | Computational fluid dynamics is a rich field and its application to the analysis of diffusion tensor imaging (DTI) datasets has yielded possible applications to tractography, image registration, and white matter pathology. We are developing several useful and novel diffusion tensor imaging (DTI) analysis algorithms modeled on the principles of fluid mechanics for inclusion within the NA-MIC framework. The goal of this project is to develop these methods, make them compatible with the NA-MIC ITK-based software infrastructure (i.e. Slicer), and promote their dissemination to the scientific community. | ||
| + | |||
| + | See our [[hageman:NAMICFluidMechDTITractography|Project Page]] for more information. | ||
| + | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| Line 32: | Line 35: | ||
====January 2008 Project Week==== | ====January 2008 Project Week==== | ||
| − | * | + | * Integration of method into Slicer 3 |
| − | * | + | ** Finished initial coding of method in itk |
| + | ** Continuing work on the following features: | ||
| + | *** Multi-Threaded Implementation | ||
| + | *** Fluid velocity vector field animation | ||
| + | *** Layout for Slicer 3 Plug-in | ||
| + | * Analysis of tractography group test data set | ||
| + | ** Finished analysis of MIND data set, M87101083_visit1, and uploaded results to database | ||
| + | ** Continuing analysis of other MIND data sets for cross-method validation | ||
| + | ** Continuing work on digital DTI phantom creation and application of method to it. For more details on the helix phantom, see our separate [[Hageman:NAMICHelixPhantom|project page]]. | ||
====June 2007 Project Week==== | ====June 2007 Project Week==== | ||
Latest revision as of 19:18, 21 February 2008
Home < 2008 Winter Project Week:Fluid Mechanics Tractography Return to 2008_Winter_Project_Week |
Key Investigators
- UCLA: Nathan Hageman
- UCLA: Arthur Toga, Ph.D
- Georgia Tech: John Melonakos (interested collaborator)
Objective
Computational fluid dynamics is a rich field and its application to the analysis of diffusion tensor imaging (DTI) datasets has yielded possible applications to tractography, image registration, and white matter pathology. We are developing several useful and novel diffusion tensor imaging (DTI) analysis algorithms modeled on the principles of fluid mechanics for inclusion within the NA-MIC framework. The goal of this project is to develop these methods, make them compatible with the NA-MIC ITK-based software infrastructure (i.e. Slicer), and promote their dissemination to the scientific community.
See our Project Page for more information.
Approach / Plan
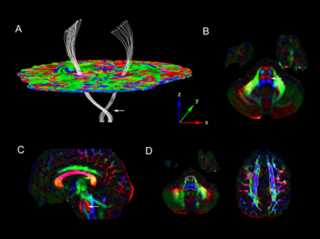
We have developed and initially validated a DTI tractography method based on Navier-Stokes fluid mechanics. See the papers listed in the reference section for complete details on the method. Our approach for this project week will focus on the following:
- Working with NA-MIC members and collaborators to successfully integrate our method in a NA-MIC ITK compatible form with Slicer.
- Working with the tractography group in evaluating data and validating our method.
In addition, we hope that our introdution of this method to the NA-MIC community will promote fruitful collaborations.
Progress
January 2008 Project Week
- Integration of method into Slicer 3
- Finished initial coding of method in itk
- Continuing work on the following features:
- Multi-Threaded Implementation
- Fluid velocity vector field animation
- Layout for Slicer 3 Plug-in
- Analysis of tractography group test data set
- Finished analysis of MIND data set, M87101083_visit1, and uploaded results to database
- Continuing analysis of other MIND data sets for cross-method validation
- Continuing work on digital DTI phantom creation and application of method to it. For more details on the helix phantom, see our separate project page.
June 2007 Project Week
We have made progress in converting our DTI analysis code to work in the ITK/vTK environment. We have created modules for
- reconstruction of the diffusion tensor and computation of common DTI scalar volumes (FA, LI, RGB)
- computation of fluid velocity vector field volume
- reconstruction of tracts based on the above fluid velocity volume
Current work is focusing on optimizing the code for the above modules and integrating them as tools in NAMIC's Slicer software environment.
References
- Hageman NS, Shattuck DW, Narr K, Toga AW (2006). A diffusion tensor imaging tractography method based on Navier-Stokes fluid mechanics. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006), Arlington, VA, USA, 6-9 April 2006. p. 798-801.
- Hageman NS, Toga AW, Narr K, Shattuck DW (2008). A diffusion tensor imaging tractography algorithm based on Navier-Stokes fluid mechanics. IEEE Trans. in Medicial Imaging, In Submission.
- Hamilton L, Nuechterlein K, Hageman NS, Woods R, Asarnow R, Alger J, Gaser C, Toga AW, Narr K (2008). Mean Diffusivity and Fractional Anisotropy as Indicators of Schizophrenia and Genetic Vulnerability, Neuroimage, In Submission.