Difference between revisions of "2008 Winter Project Week:microslicer 3"
| (5 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2008_Winter_Project_Week]] ]] | |[[Image:NAMIC-SLC.jpg|thumb|320px|Return to [[2008_Winter_Project_Week]] ]] | ||
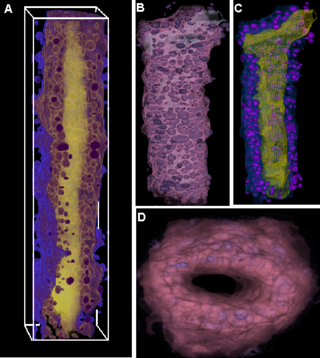
| − | |valign="top"|[[Image: | + | |valign="top"|[[Image:MammaryDuctOfMouse.png |thumb|320px|Mammary Duct of Mouse ]] |
|} | |} | ||
| Line 8: | Line 8: | ||
* The Ohio State University: Kishore Mosaliganti, Raghu Machiraju | * The Ohio State University: Kishore Mosaliganti, Raghu Machiraju | ||
* Kitware: Brad Davis, Stephen Aylward | * Kitware: Brad Davis, Stephen Aylward | ||
| − | * | + | * BWH: Steve Pieper |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 15: | Line 15: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | |||
| − | + | We have developed techniques to achieve cell/nuclei segmentations in microscopic images. Often, nuclei and cells appear as overlapping structures and splitting them apart is a non-trivial problem in image analysis. The microscopy modalities of interest include light, confocal and phase-contrast microscopy. | |
| − | + | We have also work on characterizing biological micro-structure in terms of micro-components. This helps us to perform tissue segmentations, clonal population segmentation and tracking. Once again, we propose to incorporate such techniques borrowed from material science and spatial statistics into the microscopy image analysis workflows. | |
| + | |||
| + | The objectives of this project are to port the ITK-based cell segmentation and micro-structure characterization code to Slicer3 framework. | ||
</div> | </div> | ||
| Line 32: | Line 33: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| − | + | We have currently implemented the cell segmentation algorithm using Geodesics Active Contours and Image-based Voronoi Tessellations in the ITK framework. We have also implemented the microstructure characterization algorithms using the N-Point Correlation functions in ITK. Currently, we are working on building a cell shape model to use in cell segmentations. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 47: | Line 42: | ||
===References=== | ===References=== | ||
| − | * | + | * Tensor Classification of N-point Correlation Function features for Histology Tissue Segmentation. K. Mosaliganti, F. Janoos, O. Irfanoglu, R. Ridgway, R. Machiraju, K. Huang, J. Saltz, Gustavo Leone and M. Ostrowski. In review for the Special Issue on Medical Image Analysis with Applications in Biology, Journal of Medical Image Analysis. |
| − | * | + | * Visualization of Cellular Biology Structures from Optical Microscopy Data. K. Mosaliganti, L. Cooper, R. Sharp, R. Machiraju, K. Huang and Gustavo Leone. In review at the IEEE Transactions in Visualization and Computer Graphics. |
| − | * | + | * Geometry-driven Visualization of Microscopic Structures in Biology. K. Mosaliganti, R. Machiraju, K. Huang and Gustavo Leone. In review at the Workshop on Knowledge-Assisted Visualization, IEEE Visualization Conference, Sacramento, California, 2007. |
| − | * J. | + | * Automated Quantification of Colony Growth in Clonogenic Assays. K. Mosaliganti, J. Chen, F. Janoos, R. Machiraju, W. Xia, X. Xu, K. Huang. Workshop on Medical Image Analysis with Applications in Biology, 2007, Piscatway, Rutgers, New Jersey, USA. |
| − | * | + | * Histology Image Segmentation using the N-Point Correlation Functions. F. Janoos, O. Irfanoglu, K. Mosaliganti, R. Machiraju, K. Huang, P.Wenzel, A. de Bruin, G. Leone. In Proceedings of International Symposium of Biomedical Imaging (ISBI) 2007, Washington DC, USA. |
| − | |||
Latest revision as of 02:32, 27 December 2007
Home < 2008 Winter Project Week:microslicer 3 Return to 2008_Winter_Project_Week |
Key Investigators
- The Ohio State University: Kishore Mosaliganti, Raghu Machiraju
- Kitware: Brad Davis, Stephen Aylward
- BWH: Steve Pieper
Objective
We have developed techniques to achieve cell/nuclei segmentations in microscopic images. Often, nuclei and cells appear as overlapping structures and splitting them apart is a non-trivial problem in image analysis. The microscopy modalities of interest include light, confocal and phase-contrast microscopy.
We have also work on characterizing biological micro-structure in terms of micro-components. This helps us to perform tissue segmentations, clonal population segmentation and tracking. Once again, we propose to incorporate such techniques borrowed from material science and spatial statistics into the microscopy image analysis workflows.
The objectives of this project are to port the ITK-based cell segmentation and micro-structure characterization code to Slicer3 framework.
Approach, Plan
Our approach is based upon using image tessellations and micro-structure characterization algorithms widely used in the material science community. These algorithms help us understand biological organization in terms of component packing densities, arrangements and spatial distributions. The algorithms are detailed in the references provided below. Our main purpose at the Project Week is to work with our collaborators in order to define the scope of this project, decide upon the details of the microSlicer framework in Slicer3, and implement our current developed code into the Slicer3 framework.
Progress
We have currently implemented the cell segmentation algorithm using Geodesics Active Contours and Image-based Voronoi Tessellations in the ITK framework. We have also implemented the microstructure characterization algorithms using the N-Point Correlation functions in ITK. Currently, we are working on building a cell shape model to use in cell segmentations.
References
- Tensor Classification of N-point Correlation Function features for Histology Tissue Segmentation. K. Mosaliganti, F. Janoos, O. Irfanoglu, R. Ridgway, R. Machiraju, K. Huang, J. Saltz, Gustavo Leone and M. Ostrowski. In review for the Special Issue on Medical Image Analysis with Applications in Biology, Journal of Medical Image Analysis.
- Visualization of Cellular Biology Structures from Optical Microscopy Data. K. Mosaliganti, L. Cooper, R. Sharp, R. Machiraju, K. Huang and Gustavo Leone. In review at the IEEE Transactions in Visualization and Computer Graphics.
- Geometry-driven Visualization of Microscopic Structures in Biology. K. Mosaliganti, R. Machiraju, K. Huang and Gustavo Leone. In review at the Workshop on Knowledge-Assisted Visualization, IEEE Visualization Conference, Sacramento, California, 2007.
- Automated Quantification of Colony Growth in Clonogenic Assays. K. Mosaliganti, J. Chen, F. Janoos, R. Machiraju, W. Xia, X. Xu, K. Huang. Workshop on Medical Image Analysis with Applications in Biology, 2007, Piscatway, Rutgers, New Jersey, USA.
- Histology Image Segmentation using the N-Point Correlation Functions. F. Janoos, O. Irfanoglu, K. Mosaliganti, R. Machiraju, K. Huang, P.Wenzel, A. de Bruin, G. Leone. In Proceedings of International Symposium of Biomedical Imaging (ISBI) 2007, Washington DC, USA.