Difference between revisions of "2009 Summer Project Week-FastMarching for brain tumor segmentation"
| Line 4: | Line 4: | ||
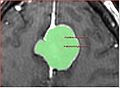
Image:fastmarching1.jpg|Falcine meningioma segmentation | Image:fastmarching1.jpg|Falcine meningioma segmentation | ||
Image:fastmarching2.jpg|Convexity meningioma segmentation | Image:fastmarching2.jpg|Convexity meningioma segmentation | ||
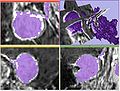
| + | Image:fastmarching3.jpg|Problem illustrated | ||
</gallery> | </gallery> | ||
| Line 22: | Line 23: | ||
<h3>Approach, plan</h3> | <h3>Approach, plan</h3> | ||
We will evaluate the fast marching segmentation methods developed by the GeorgiaTech collaborators on clinical meningioma MRI data. A set of anonymized images representing meningiomas of various appearance will be used for testing. Based on the results of the evaluation, decisions will be made about the most appropriate method to be implemented as a Slicer module. | We will evaluate the fast marching segmentation methods developed by the GeorgiaTech collaborators on clinical meningioma MRI data. A set of anonymized images representing meningiomas of various appearance will be used for testing. Based on the results of the evaluation, decisions will be made about the most appropriate method to be implemented as a Slicer module. | ||
| + | |||
| + | The major issue with the fast marching segmentation available in Slicer3 currently is that it leaks before it conforms to the boundary of the structure. We would like to investigate if this issue can be remedied. | ||
</div> | </div> | ||
Revision as of 14:48, 17 June 2009
Home < 2009 Summer Project Week-FastMarching for brain tumor segmentationKey Investigators
- BWH: Andriy Fedorov, Ron Kikinis
- GeorgiaTech: Yi Gao
Objective
We will investigate the currently available algorithms and tools for fast marching segmentation, and study their applicability to the problem of segmenting meningiomas in contrast-enhanced brain MRI.
The key requirements to the segmentation approach are naturally accuracy, but also simplicity of initialization and acceptable speed for interactive analysis.
Approach, plan
We will evaluate the fast marching segmentation methods developed by the GeorgiaTech collaborators on clinical meningioma MRI data. A set of anonymized images representing meningiomas of various appearance will be used for testing. Based on the results of the evaluation, decisions will be made about the most appropriate method to be implemented as a Slicer module.
The major issue with the fast marching segmentation available in Slicer3 currently is that it leaks before it conforms to the boundary of the structure. We would like to investigate if this issue can be remedied.
Progress
FastMarchingSegmentation module by Eric Pichon was ported into Slicer3.