Difference between revisions of "2009 Summer Project Week 4D Gated US In Slicer"
(Created page with '__NOTOC__ <gallery> Image:PW2009-v3.png|Project Week Main Page Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus ...') |
|||
| (22 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Project Week Main Page]] |
| − | Image: | + | Image:3DUSReconstructionSchematic.png|Schematic of 3D ultrasound reconstruction |
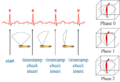
| − | Image: | + | Image:GatedUSReconstructionSchematic.png|Schematic of gated (4D) ultrasound reconstruction |
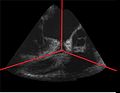
| + | Image:GatedUSReconstruction rotationalTEE_0R-R.jpg|Example reconstructed ultrasound volume of a porcine heart, at 0% R-R | ||
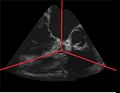
| + | Image:GatedUSReconstruction rotationalTEE_50R-R.jpg|Example reconstructed ultrasound volume of a porcine heart, at 50% R-R | ||
| + | |||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Robarts Research Institute / University of Western Ontario: Danielle Pace |
| − | |||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | The objective of this project is to enable gated four-dimensional ultrasound reconstruction in Slicer3. | |
| − | |||
| − | |||
| − | |||
| + | We have previously written and validated software that performs 4D ultrasound reconstruction from a tracked 2D probe in real-time (Pace et al, SPIE 2009). This allows the user to interact with the reconstructed time series of 3D volumes as they are being acquired and to adjust the acquisition process as necessary. The user-interface for the reconstruction software has been implemented within the [http://www.atamai.com Atamai Viewer] framework. During the project week, we will integrate it into Slicer3. | ||
| Line 34: | Line 27: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | We plan to interface the reconstruction software with Slicer3 using the SynchroGrab framework described by (Boisvert et al, MICCAI 2008). A command-line interface and a calibration file will allow the user to specify the reconstruction parameters. While the gated reconstruction is performed, the resulting time series of volumes will be transferred to Slicer3 using the OpenIGTLink protocol for interactive visualization. | |
| − | |||
</div> | </div> | ||
| Line 43: | Line 35: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Discussions with Noby Hata pointed us to an extension of the SynchroGrab code base previously written by Jan Gumprecht, and so we decided to use this as our starting point. | |
| − | + | * During the project week, we began modifying this software to work with our real-time 4D US reconstruction classes. | |
| + | * To visualize the 4D dataset within Slicer3, Junichi Tokuda's [http://www.na-mic.org/Wiki/index.php/2009_Summer_Project_Week_4D_Imaging 4D imaging module] will be used to toggle between the different 3D volumes and to play a cine loop of the entire dataset. | ||
| + | * Further work includes testing the software in the lab with actual ultrasound and tracking devices. | ||
</div> | </div> | ||
| Line 52: | Line 46: | ||
==References== | ==References== | ||
| − | * | + | *Pace DF, Wiles AD, Moore J, Wedlake C, Gobbi DG, Peters TM, [http://spie.org/x648.html?product_id=813705 Validation of four-dimensional ultrasound for targetting in minimally-invasive beating-heart surgery], Proceedings of SPIE Medical Imaging: Visualization, Image-Guided Procedures and Modeling, 2009. |
| − | * | + | *Boisvert J, Gobbi DG, Vikal S, Rohling R, Fichtinger G, Abolmaesumi P, [http://www.midasjournal.org/browse/publication/618 An open-source solution for interactive acquisition, processing and transfer of interventional ultrasound images], Workshop on Systems and Architectures for Computer Assisted Interventions, MICCAI 2008. |
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 13:27, 26 June 2009
Home < 2009 Summer Project Week 4D Gated US In SlicerKey Investigators
- Robarts Research Institute / University of Western Ontario: Danielle Pace
Objective
The objective of this project is to enable gated four-dimensional ultrasound reconstruction in Slicer3.
We have previously written and validated software that performs 4D ultrasound reconstruction from a tracked 2D probe in real-time (Pace et al, SPIE 2009). This allows the user to interact with the reconstructed time series of 3D volumes as they are being acquired and to adjust the acquisition process as necessary. The user-interface for the reconstruction software has been implemented within the Atamai Viewer framework. During the project week, we will integrate it into Slicer3.
Approach, Plan
We plan to interface the reconstruction software with Slicer3 using the SynchroGrab framework described by (Boisvert et al, MICCAI 2008). A command-line interface and a calibration file will allow the user to specify the reconstruction parameters. While the gated reconstruction is performed, the resulting time series of volumes will be transferred to Slicer3 using the OpenIGTLink protocol for interactive visualization.
Progress
- Discussions with Noby Hata pointed us to an extension of the SynchroGrab code base previously written by Jan Gumprecht, and so we decided to use this as our starting point.
- During the project week, we began modifying this software to work with our real-time 4D US reconstruction classes.
- To visualize the 4D dataset within Slicer3, Junichi Tokuda's 4D imaging module will be used to toggle between the different 3D volumes and to play a cine loop of the entire dataset.
- Further work includes testing the software in the lab with actual ultrasound and tracking devices.
References
- Pace DF, Wiles AD, Moore J, Wedlake C, Gobbi DG, Peters TM, Validation of four-dimensional ultrasound for targetting in minimally-invasive beating-heart surgery, Proceedings of SPIE Medical Imaging: Visualization, Image-Guided Procedures and Modeling, 2009.
- Boisvert J, Gobbi DG, Vikal S, Rohling R, Fichtinger G, Abolmaesumi P, An open-source solution for interactive acquisition, processing and transfer of interventional ultrasound images, Workshop on Systems and Architectures for Computer Assisted Interventions, MICCAI 2008.