Difference between revisions of "2009 Summer Project Week Hageman DTIDigitalPhantom"
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{| | {| | ||
| − | |[[Image:PW2009-v3.png|thumb|320px|[[2009_Summer_Project_Week|Project Week Main Page]] ]] | + | |[[Image:PW2009-v3.png|thumb|320px|[[2009_Summer_Project_Week#Projects|Project Week Main Page]] ]] |
|[[Image:Hageman_DWIPhantomWebPage4NAMIC_09-06-13.jpg|thumb|320px|Web service for DWI phantom generator hosted by UCLA]] | |[[Image:Hageman_DWIPhantomWebPage4NAMIC_09-06-13.jpg|thumb|320px|Web service for DWI phantom generator hosted by UCLA]] | ||
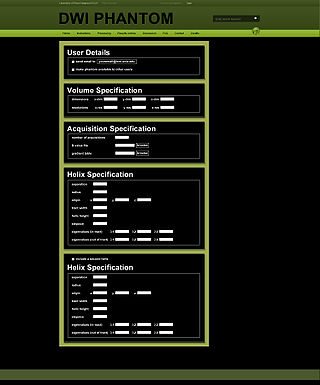
| + | |[[Image:Hageman_DWIPhantomForm4NAMIC_09-06-13.jpg|thumb|320px|Form used by web service to generate phantom DWI dataset]] | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| Line 20: | Line 18: | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
| − | + | Imaging datasets constructed from digital phantom models continue to be valuable resource for validating image analysis algorithms. There is a need for such resources for the validation of DTI analysis algorithms, especially for those methods that involve measures of WM integrity or segmentation of WM tracts. Although some fixed digital DTI phantom models exist, we have created a software tool that allows users to generate a DWI dataset from a ground truth WM shape based on their own chosen experimental paradigm. This will provide users with the flexibility to define the parameters of each validation dataset, which is not available with current digital DTI phantom validation resources. | |
| + | |||
| + | The current version of the program that will generate a DWI dataset based on a single or double helix ground truth that simulates a white matter tract. The user is free to specify desired dimensions, experimental parameters, and level of Rician or Gaussian noise in the final dataset. We will host this program via a webservice from the Laboratory of Neuroimaging at UCLA. | ||
</div> | </div> | ||
| Line 26: | Line 26: | ||
<h1>Approach, Plan</h1> | <h1>Approach, Plan</h1> | ||
| + | The functioning webservice and binaries are scheduled for an external release in July 2009. For this project week, | ||
| + | *We will demo the program and webservice utility or generate specific datasets for any interested groups. Pre-release binaries are available upon request. | ||
| + | *We will discuss possible links to the webservice as part of Slicer and NAMIC Wiki. | ||
</div> | </div> | ||
| Line 32: | Line 35: | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| − | + | *Interested collaborators met concerning release date of web service and binaries. | |
| + | *Discussion about incorporation of web service into Slicer and NAMIC Wiki. | ||
</div> | </div> | ||
| Line 41: | Line 45: | ||
===References=== | ===References=== | ||
* Hageman NS, Shattuck DW, Narr K, Toga AW (2006). A diffusion tensor imaging tractography method based on Navier-Stokes fluid mechanics. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006), Arlington, VA, USA, 6-9 April 2006. p. 798-801 | * Hageman NS, Shattuck DW, Narr K, Toga AW (2006). A diffusion tensor imaging tractography method based on Navier-Stokes fluid mechanics. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006), Arlington, VA, USA, 6-9 April 2006. p. 798-801 | ||
| − | * Hageman NS, Toga AW, Narr K, Shattuck DW ( | + | * Hageman NS, Toga AW, Narr K, Shattuck DW (2009). A diffusion tensor imaging tractography algorithm based on Navier-Stokes fluid mechanics. IEEE Trans. in Medicial Imaging 28(3): 348-360. |
Latest revision as of 14:12, 13 July 2009
Home < 2009 Summer Project Week Hageman DTIDigitalPhantom
Key Investigators
- UCLA: Nathan Hageman
- UCLA: Arthur Toga, Ph.D
Objective
Imaging datasets constructed from digital phantom models continue to be valuable resource for validating image analysis algorithms. There is a need for such resources for the validation of DTI analysis algorithms, especially for those methods that involve measures of WM integrity or segmentation of WM tracts. Although some fixed digital DTI phantom models exist, we have created a software tool that allows users to generate a DWI dataset from a ground truth WM shape based on their own chosen experimental paradigm. This will provide users with the flexibility to define the parameters of each validation dataset, which is not available with current digital DTI phantom validation resources.
The current version of the program that will generate a DWI dataset based on a single or double helix ground truth that simulates a white matter tract. The user is free to specify desired dimensions, experimental parameters, and level of Rician or Gaussian noise in the final dataset. We will host this program via a webservice from the Laboratory of Neuroimaging at UCLA.
Approach, Plan
The functioning webservice and binaries are scheduled for an external release in July 2009. For this project week,
- We will demo the program and webservice utility or generate specific datasets for any interested groups. Pre-release binaries are available upon request.
- We will discuss possible links to the webservice as part of Slicer and NAMIC Wiki.
Progress
- Interested collaborators met concerning release date of web service and binaries.
- Discussion about incorporation of web service into Slicer and NAMIC Wiki.
References
- Hageman NS, Shattuck DW, Narr K, Toga AW (2006). A diffusion tensor imaging tractography method based on Navier-Stokes fluid mechanics. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006), Arlington, VA, USA, 6-9 April 2006. p. 798-801
- Hageman NS, Toga AW, Narr K, Shattuck DW (2009). A diffusion tensor imaging tractography algorithm based on Navier-Stokes fluid mechanics. IEEE Trans. in Medicial Imaging 28(3): 348-360.