Difference between revisions of "2009 Summer Project Week Meningioma growth simulation"
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] |
Image:tumorsim-meningioma1.jpg|Tumor sumulation iteration 1 | Image:tumorsim-meningioma1.jpg|Tumor sumulation iteration 1 | ||
Image:tumorsim-meningioma2.jpg|... iteration 2 | Image:tumorsim-meningioma2.jpg|... iteration 2 | ||
Image:tumorsim-meningioma3.jpg|... iteration 3 | Image:tumorsim-meningioma3.jpg|... iteration 3 | ||
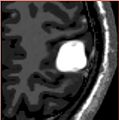
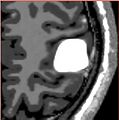
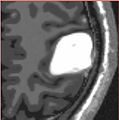
| + | |Iterations 1, 3 and 10 of the simulation with increased stiffness and identified parameters | ||
| + | Image:meningiomasim_iter1.jpg| | ||
| + | Image:meningiomasim_iter3.jpg| | ||
| + | Image:meningiomasim_iter10.jpg| | ||
</gallery> | </gallery> | ||
| Line 15: | Line 19: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Meningiomas are primary brain tumors that grow from the cells of meninges. Such tumors are typically benign, and are not invasive. In this project we will investigate the use of the tumor growth simulation tool, [http://www.ucnia.org/softwaredata/5-tumordata/10-simtumordb.html TumorSim], for the purposes of modeling the growth of meningioma. Our primary objective is to tune the parameters of the simulation to have maximally realistic appearance of contrast-enhanced tumor mass, realistic deformation of the surrounding tissue, and to evaluate the capabilities of the tumor simulator in modeling meningiomas of various morphology. | |
| − | |||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, plan</h3> | <h3>Approach, plan</h3> | ||
| + | TumorSim software allows to specify the following simulation parameters: | ||
| + | * deformation parameters | ||
| + | ** number of iterations | ||
| + | ** initial pressure | ||
| + | ** kappa | ||
| + | * infiltration parameters | ||
| + | ** number of iterations | ||
| + | ** number of body force iterations | ||
| + | ** time step | ||
| + | ** body force coefficient | ||
| + | * contrast enhancement type: none, ring or uniform | ||
| + | |||
| + | We will analyze contrast-enhanced MRIs of meningiomas of different sizes, located in different areas of brain, and will attempt to develop a set of recommendations for initializations of the parameters to mimic these tumors. | ||
| + | |||
| + | Additional practical considerations: | ||
| + | * how can the level of noise be controlled? | ||
| + | * what is the influence of the initialization label size on the tumor shape? | ||
| + | * tumor-brain interface appearance and correspondence with the tumor label | ||
| + | |||
</div> | </div> | ||
<div style="width: 40%; float: left;"> | <div style="width: 40%; float: left;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| + | * initial set of parameters that characterize meningioma MR appearance were suggested | ||
| + | * simulated images were generated using different iteration values for initial assessment | ||
| + | * TumorSim has been augmented with more simulation parameters exposed, which allows more realism into simulation results | ||
| + | * direction to be explored: improving mesh used in the simulation (collaboration with Ross Whitaker) | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | <h3>Meningioma simulation parameter setup</h3> | ||
| + | |||
| + | The following xml configuration was used for initial evaluation: | ||
| + | <pre> | ||
| + | <?xml version="1.0"?> | ||
| + | <tumor-simulation-parameters> | ||
| + | <dataset-name>SimMeningioma001</dataset-name> | ||
| + | <input-directory>/scratch/prastawa/TumorSim/TumorSimInput</input-directory> | ||
| + | <output-directory>/scratch/prastawa/TumorSim/Meningioma001</output-directory> | ||
| + | <deformation-seed>/scratch/prastawa/TumorSim/seed_m1.mha</deformation-seed> | ||
| + | <deformation-iterations>8</deformation-iterations> | ||
| + | <infiltration-iterations>4</infiltration-iterations> | ||
| + | <infiltration-body-force-iterations>4</infiltration-body-force-iterations> | ||
| + | <deformation-initial-pressure>5</deformation-initial-pressure> | ||
| + | <deformation-kappa>100</deformation-kappa> | ||
| + | <infiltration-time-step>0.5</infiltration-time-step> | ||
| + | <infiltration-body-force-coefficient>8</infiltration-body-force-coefficient> | ||
| + | <contrast-enhancement-type>uniform</contrast-enhancement-type> | ||
| + | </tumor-simulation-parameters> | ||
| + | </pre> | ||
| + | |||
| + | * paths to explore suggested by Marcel: use smaller value for the initial pressure (2) and more iterations; mesh may also be a problem -- if the first doesn't work, try to use better mesh. 10 iterations took 3 hr 32 min on george.bwh.harvard.edu. | ||
| + | |||
| + | * simulation with the reduced initial pressure did not bring desired improvements in simulation realism. Next steps -- expose some of the parameters that are currently hidden: | ||
| + | ** noise for deformation direction: influences the shape of the simulated tumor | ||
| + | ** image noise: currently high and fixed, but the real clinical images are not that noisy | ||
| + | ** image contrast (not that important) | ||
| + | ** MRI field inhomogeneity -- currently fixed | ||
| + | ** dampening: decreases with the deformation iterations | ||
| + | ** solver convergence | ||
| + | ** mesh quality can be insufficient | ||
| + | |||
| + | * last iteration of parameters we came up with: | ||
| + | <pre> | ||
| + | <?xml version="1.0"?> | ||
| + | <tumor-simulation-parameters> | ||
| + | <dataset-name>SimMeningioma001</dataset-name> | ||
| + | <input-directory>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/TumorSimInput</input-directory> | ||
| + | <output-directory>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/m1_iter1_p2</output-directory> | ||
| + | <deformation-seed>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/seed_m1.mha</deformation-seed> | ||
| + | <deformation-iterations>1</deformation-iterations> | ||
| + | <infiltration-iterations>2</infiltration-iterations> | ||
| + | <infiltration-body-force-iterations>4</infiltration-body-force-iterations> | ||
| + | <deformation-initial-pressure>1</deformation-initial-pressure> | ||
| + | <deformation-kappa>1e+06</deformation-kappa> | ||
| + | <deformation-damping>1</deformation-damping> | ||
| + | <infiltration-time-step>0.5</infiltration-time-step> | ||
| + | <infiltration-body-force-coefficient>10</infiltration-body-force-coefficient> | ||
| + | <contrast-enhancement-type>uniform</contrast-enhancement-type> | ||
| + | <brain-young-modulus>694</brain-young-modulus> | ||
| + | <brain-poisson-ratio>0.5</brain-poisson-ratio> | ||
| + | <gad-noise-stddev>1e-05</gad-noise-stddev> | ||
| + | <t1-noise-stddev>1e-05</t1-noise-stddev> | ||
| + | <t2-noise-stddev>1e-05</t2-noise-stddev> | ||
| + | <gad-max-bias-degree>0</gad-max-bias-degree> | ||
| + | <t1-max-bias-degree>0</t1-max-bias-degree> | ||
| + | <t2-max-bias-degree>0</t2-max-bias-degree> | ||
| + | <deformation-solver-iterations>10</deformation-solver-iterations> | ||
| + | <infiltration-solver-iterations>2</infiltration-solver-iterations> | ||
| + | </tumor-simulation-parameters> | ||
| + | </pre> | ||
| + | |||
</div> | </div> | ||
| + | |||
| + | <div style="width: 97%; float: left;"> | ||
<h3>References</h3> | <h3>References</h3> | ||
| + | * M.Prastawa, E.Bullitt, and G.Gerig. Simulation of Brain Tumors in MR Images for Evaluation of Segmentation Efficacy. Medical Image Analysis 13(2):297-311, 2009 [http://www.sci.utah.edu/~prastawa/papers/MedIAXX_Prastawa_SimTumorDRAFT.pdf] | ||
* [http://www.ucnia.org/softwaredata/5-tumordata/10-simtumordb.html TumorSim: Simulated Brain Tumor MRI Software and data] | * [http://www.ucnia.org/softwaredata/5-tumordata/10-simtumordb.html TumorSim: Simulated Brain Tumor MRI Software and data] | ||
| + | * [http://en.wikipedia.org/wiki/Meningioma Wikipedia: Meningioma] | ||
| + | |||
| + | </div> | ||
Latest revision as of 14:15, 26 June 2009
Home < 2009 Summer Project Week Meningioma growth simulationKey Investigators
- BWH: Andriy Fedorov, Ron Kikinis
- Utah: Marcel Prastawa
Objective
Meningiomas are primary brain tumors that grow from the cells of meninges. Such tumors are typically benign, and are not invasive. In this project we will investigate the use of the tumor growth simulation tool, TumorSim, for the purposes of modeling the growth of meningioma. Our primary objective is to tune the parameters of the simulation to have maximally realistic appearance of contrast-enhanced tumor mass, realistic deformation of the surrounding tissue, and to evaluate the capabilities of the tumor simulator in modeling meningiomas of various morphology.
Approach, plan
TumorSim software allows to specify the following simulation parameters:
- deformation parameters
- number of iterations
- initial pressure
- kappa
- infiltration parameters
- number of iterations
- number of body force iterations
- time step
- body force coefficient
- contrast enhancement type: none, ring or uniform
We will analyze contrast-enhanced MRIs of meningiomas of different sizes, located in different areas of brain, and will attempt to develop a set of recommendations for initializations of the parameters to mimic these tumors.
Additional practical considerations:
- how can the level of noise be controlled?
- what is the influence of the initialization label size on the tumor shape?
- tumor-brain interface appearance and correspondence with the tumor label
Progress
- initial set of parameters that characterize meningioma MR appearance were suggested
- simulated images were generated using different iteration values for initial assessment
- TumorSim has been augmented with more simulation parameters exposed, which allows more realism into simulation results
- direction to be explored: improving mesh used in the simulation (collaboration with Ross Whitaker)
Meningioma simulation parameter setup
The following xml configuration was used for initial evaluation:
<?xml version="1.0"?> <tumor-simulation-parameters> <dataset-name>SimMeningioma001</dataset-name> <input-directory>/scratch/prastawa/TumorSim/TumorSimInput</input-directory> <output-directory>/scratch/prastawa/TumorSim/Meningioma001</output-directory> <deformation-seed>/scratch/prastawa/TumorSim/seed_m1.mha</deformation-seed> <deformation-iterations>8</deformation-iterations> <infiltration-iterations>4</infiltration-iterations> <infiltration-body-force-iterations>4</infiltration-body-force-iterations> <deformation-initial-pressure>5</deformation-initial-pressure> <deformation-kappa>100</deformation-kappa> <infiltration-time-step>0.5</infiltration-time-step> <infiltration-body-force-coefficient>8</infiltration-body-force-coefficient> <contrast-enhancement-type>uniform</contrast-enhancement-type> </tumor-simulation-parameters>
- paths to explore suggested by Marcel: use smaller value for the initial pressure (2) and more iterations; mesh may also be a problem -- if the first doesn't work, try to use better mesh. 10 iterations took 3 hr 32 min on george.bwh.harvard.edu.
- simulation with the reduced initial pressure did not bring desired improvements in simulation realism. Next steps -- expose some of the parameters that are currently hidden:
- noise for deformation direction: influences the shape of the simulated tumor
- image noise: currently high and fixed, but the real clinical images are not that noisy
- image contrast (not that important)
- MRI field inhomogeneity -- currently fixed
- dampening: decreases with the deformation iterations
- solver convergence
- mesh quality can be insufficient
- last iteration of parameters we came up with:
<?xml version="1.0"?> <tumor-simulation-parameters> <dataset-name>SimMeningioma001</dataset-name> <input-directory>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/TumorSimInput</input-directory> <output-directory>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/m1_iter1_p2</output-directory> <deformation-seed>/projects/birn/fedorov/projects/BSF-Meningioma/MeningiomaSim/seed_m1.mha</deformation-seed> <deformation-iterations>1</deformation-iterations> <infiltration-iterations>2</infiltration-iterations> <infiltration-body-force-iterations>4</infiltration-body-force-iterations> <deformation-initial-pressure>1</deformation-initial-pressure> <deformation-kappa>1e+06</deformation-kappa> <deformation-damping>1</deformation-damping> <infiltration-time-step>0.5</infiltration-time-step> <infiltration-body-force-coefficient>10</infiltration-body-force-coefficient> <contrast-enhancement-type>uniform</contrast-enhancement-type> <brain-young-modulus>694</brain-young-modulus> <brain-poisson-ratio>0.5</brain-poisson-ratio> <gad-noise-stddev>1e-05</gad-noise-stddev> <t1-noise-stddev>1e-05</t1-noise-stddev> <t2-noise-stddev>1e-05</t2-noise-stddev> <gad-max-bias-degree>0</gad-max-bias-degree> <t1-max-bias-degree>0</t1-max-bias-degree> <t2-max-bias-degree>0</t2-max-bias-degree> <deformation-solver-iterations>10</deformation-solver-iterations> <infiltration-solver-iterations>2</infiltration-solver-iterations> </tumor-simulation-parameters>
References
- M.Prastawa, E.Bullitt, and G.Gerig. Simulation of Brain Tumors in MR Images for Evaluation of Segmentation Efficacy. Medical Image Analysis 13(2):297-311, 2009 [1]
- TumorSim: Simulated Brain Tumor MRI Software and data
- Wikipedia: Meningioma