Difference between revisions of "2009 Summer Project Week Slicer3 Adaptive Radiotherapy"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW2009-v3.png|Project Week Main Page Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus ...') |
|||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week|Project Week Main Page]] | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Project Week Main Page]] |
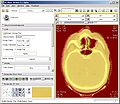
| − | Image: | + | Image:PW2009-ART-rigid.jpg|Rigid registration of pre-treatment and mid-treatment scan. |
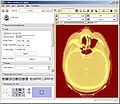
| − | Image: | + | Image:PW2009-ART-warped.jpg|Deformable registration of pre-treatment and mid-treatment scan. |
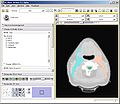
| + | Image:PW2009-ART-ss.jpg|Radiotherapy structures warped from pre-treatment to mid-treatment scan. | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * MGH: Greg Sharp, Rui Li, Annie Chan |
| − | * | + | * NA-MIC: Steve Pieper, Wendy Plesniak |
| + | * Magna Graecia University: Maria Francesca Spadea | ||
| + | * Politecnico di Milano: Marta Peroni | ||
| + | * MIT: Polina Golland | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 18: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | <ul> | |
| − | + | <li>Add DicomRT Structure Set import to labelmap</li> | |
| − | + | <li>Investigate DicomRT RTDOSE import</li> | |
| − | + | <li>Check RTSS and RTDOSE storage in XNAT (Dan Marcus)</li> | |
| − | + | <li>Discuss long-term solution for DicomRT sturctures (labelmaps are not suitable for overlapping structures)</li> | |
| − | + | <li>Discuss Dicom RTDOSE import and display</li> | |
| + | <li>Discuss source code sharing strategy</li> | ||
| + | </ul> | ||
</div> | </div> | ||
| Line 33: | Line 32: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | <ul> | |
| − | + | <li>Discuss strategy.</li> | |
| − | + | </ul> | |
| − | |||
| − | |||
</div> | </div> | ||
| Line 43: | Line 40: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | <ul> | |
| − | + | <li>Discuss and review: mesh representation using VTK File I/O.</li> | |
| − | + | <li> | |
| + | Upload DicomRT sample to wiki: [[media:Xio-4.33-chest_phantom_dicomrt.tar.gz]] | ||
| + | </li> | ||
| + | </ul> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
==References== | ==References== | ||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 15:31, 26 June 2009
Home < 2009 Summer Project Week Slicer3 Adaptive RadiotherapyKey Investigators
- MGH: Greg Sharp, Rui Li, Annie Chan
- NA-MIC: Steve Pieper, Wendy Plesniak
- Magna Graecia University: Maria Francesca Spadea
- Politecnico di Milano: Marta Peroni
- MIT: Polina Golland
Objective
- Add DicomRT Structure Set import to labelmap
- Investigate DicomRT RTDOSE import
- Check RTSS and RTDOSE storage in XNAT (Dan Marcus)
- Discuss long-term solution for DicomRT sturctures (labelmaps are not suitable for overlapping structures)
- Discuss Dicom RTDOSE import and display
- Discuss source code sharing strategy
Approach, Plan
- Discuss strategy.
Progress
- Discuss and review: mesh representation using VTK File I/O.
- Upload DicomRT sample to wiki: media:Xio-4.33-chest_phantom_dicomrt.tar.gz