Difference between revisions of "2009 Summer Project Week Slicer3 XNAT usecases"
| Line 58: | Line 58: | ||
'''Step4.''' Researcher queries for case data & browses results using GUI, downloads to local disk for study. | '''Step4.''' Researcher queries for case data & browses results using GUI, downloads to local disk for study. | ||
| − | == | + | == GUI Requirements == |
* Interface to commercial systems? | * Interface to commercial systems? | ||
* Appropriateness of XND for tagging & upload of non-imaging assessors? | * Appropriateness of XND for tagging & upload of non-imaging assessors? | ||
* Specific user requirements for metadata authoring and upload client software? | * Specific user requirements for metadata authoring and upload client software? | ||
| − | == | + | == Webservices API Requirements |
| + | |||
| + | == Metadata Requirements or Data Model modifications == | ||
* DCMRT supported by DICOM rule? | * DCMRT supported by DICOM rule? | ||
Revision as of 12:10, 24 June 2009
Home < 2009 Summer Project Week Slicer3 XNAT usecasesContents
XNAT interface Worksheet
Goals:
- Determine User Interface Requirements for representative use cases
- Determine Webservices API Requirements for representative use cases
- Determine sufficiency of XNAT Data Model for representative use cases
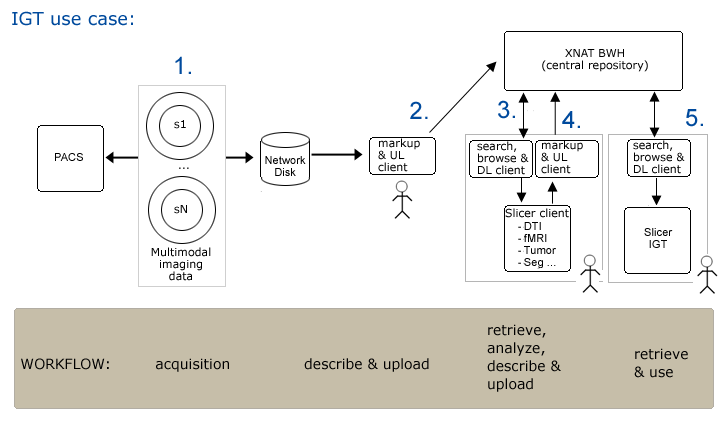
Use Case 1: Image Guided Therapy Planning and in OR
Workflow
Step1. Subject is scanned and data is pushed to a networked disk (and referenced by a PACS system) automatically.
Step2. Researcher1 uses an XND client (on a laptop or a desktop system) to
- apply metadata (DICOM rule sufficient?)
- upload data to XNAT repository
Step3. Researcher2 opens slicer and querieswebservices for a list of subjects.
- Query for all subjects on system
- Choose a subject, query for available data...
- Browse results
- download data of interest
Step4. Completed neurosurgical planning analysis
- apply metadata (need custom metadata) --must be quick/easy to do
- and upload results -- must be quick/easy to do (drag/drop, with confirmation0).
Step5. In IGT, search for and download scene for patient (subject)
Clinician cuts network link to remote repository (all files are locally cached so OR system is self sufficient).
GUI Requirements
== Webservices API Requirements
Metadata Requirements or Data Model modifications
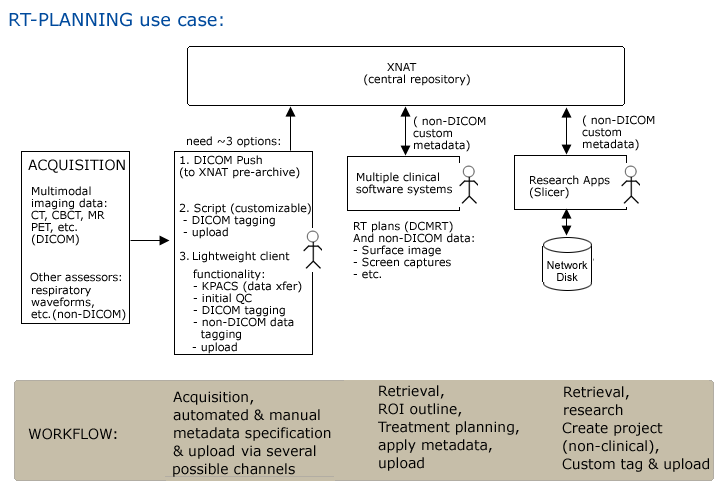
Use Case 2: Radiation Treatment Planning
Workflow
Step1. Patient is scanned and data is pushed to XNAT (and referenced by a PACS system) automatically.
- possibly DICOM DCMRT metadata automatically applied
- other assessors may be collected and described manually, uploaded to XNAT using client GUI.
Step2. Scientist/Clinician queries for patient data & browses results using GUI, downloads to RT planning software
Step3. Scientist/Clinician uploads plan in progress (drag & drop?). DICOM DCMRT metadata automatically applied at upload.
Step4. Researcher queries for case data & browses results using GUI, downloads to local disk for study.
GUI Requirements
- Interface to commercial systems?
- Appropriateness of XND for tagging & upload of non-imaging assessors?
- Specific user requirements for metadata authoring and upload client software?
== Webservices API Requirements
Metadata Requirements or Data Model modifications
- DCMRT supported by DICOM rule?
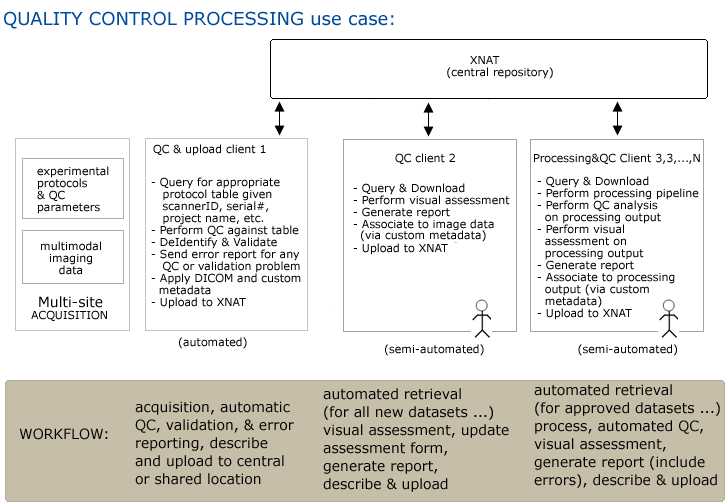
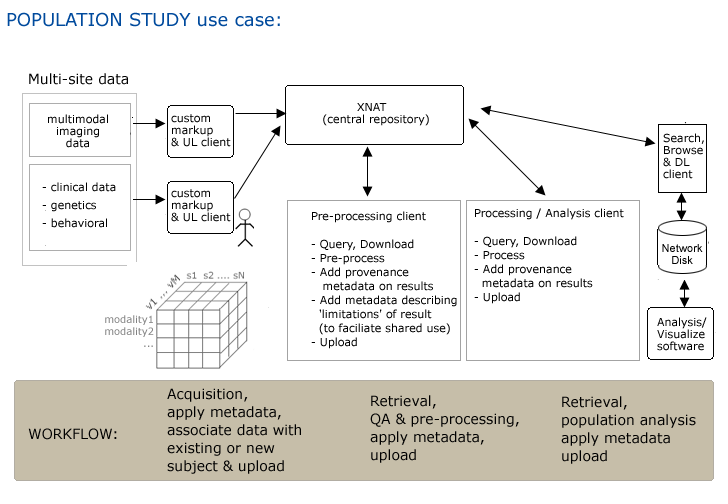
Use Case 3: Quality Control Processing of Large Studies
Workflow
Step 1. Multiple study sites acquire imaging data and other assessors
Step 2. Combination of automatic (DICOM) and manual (custom?) markup of data and upload to central repository
Step 3. QC batch client 1 automatically queries for (site/subject/scanID), retrieves, processes data, generates report and notifies any site of errors in (or missing) datasets. Additional processing may be performed (de-identification of data)...
Step4. Additional metadata may be automatically applied (provenance?) and data is uploaded.
Step5. QC batch client 2 (automatically?) queries for (site/subject/scanID), retrieves, allows visual inspection of data and report generation.
Step6. Report is automatically uploaded and associated with image data.
Specific Interface Requirements
Specific Metadata Requirements or Data Model modifications
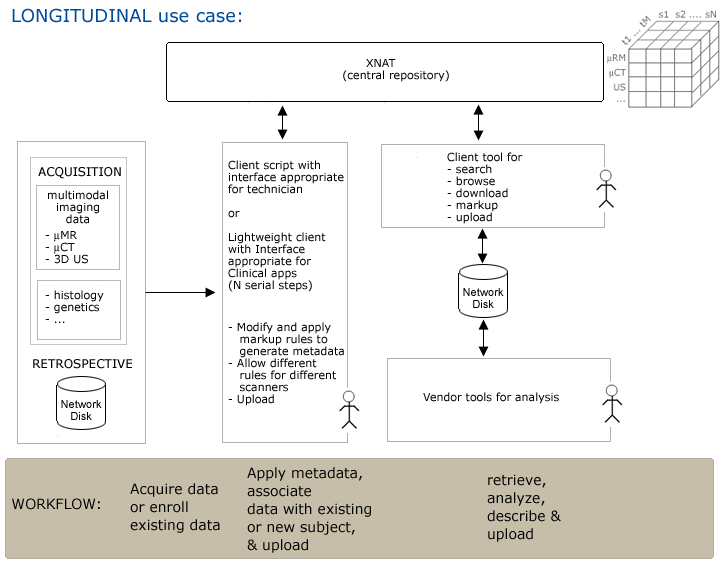
Use Case 4: Longitudinal Studies
Workflow
Step1. Imaging data and other assessors (induced pathology, etc.) are acquired and saved to a network disk.
Step2. Research assistant uses GUI to apply metadata at capture, or subsequently & uploads to XNAT.
- Need to specify whether subject is control or experimental
- Imagedata is typically in non-standard DICOM format.
- Need to associate new data to existing (or new) subject in longitudinal study -- need entities for:
- modality
- subject
- timepoint
- ...?
Step3. Researcher queries for data using GUI, browses results and downloads to vendor tools for analysis.
Step4. Researcher generates derived data,
- applies metadata (provenance? and maybe notable parameters of interest).
- associates to subject or study (population)
- and uploads via GUI
Specific Interface Requirements
Specific Metadata Requirements or Data Model modifications