Difference between revisions of "2009 Winter Project Week ChangeTracker"
| Line 66: | Line 66: | ||
===Volume rendering of label data=== | ===Volume rendering of label data=== | ||
| + | {| | ||
| + | | | ||
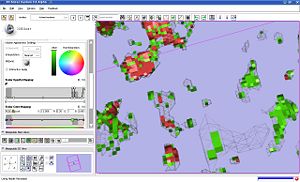
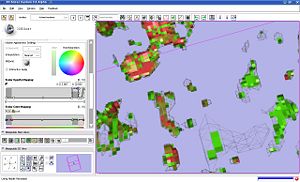
We investigated the behavior of volume rendering on the sample data from tumor growth analysis. We identified that it is crucial to use nearest neighbor interpolation for label volume rendering. It appears that the behavior of the opacity mapping function is rather unexplained (see the screenshots): | We investigated the behavior of volume rendering on the sample data from tumor growth analysis. We identified that it is crucial to use nearest neighbor interpolation for label volume rendering. It appears that the behavior of the opacity mapping function is rather unexplained (see the screenshots): | ||
* although the input label map contains only three intensity values (background 0, shrinking 12 and growth 14), the cut-off location of the opacity mapping for the shrinking label affects rendering | * although the input label map contains only three intensity values (background 0, shrinking 12 and growth 14), the cut-off location of the opacity mapping for the shrinking label affects rendering | ||
* the results of volume rendering are drastically different from the surface recovered with marching cubes (see the screenshots) | * the results of volume rendering are drastically different from the surface recovered with marching cubes (see the screenshots) | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Programmatic initialization of the opacity transfer function for two-label data: | Programmatic initialization of the opacity transfer function for two-label data: | ||
| Line 100: | Line 97: | ||
''Current solutions/workarounds we consider: use surface rendering from Marching cubes instead of volume rendering.'' | ''Current solutions/workarounds we consider: use surface rendering from Marching cubes instead of volume rendering.'' | ||
| + | |||
| + | | | ||
| + | {| | ||
| + | |- | ||
| + | |[[Image:transfer_function1.jpg|thumb|300px|Initial transfer function]] | ||
| + | |- | ||
| + | |[[Image:transfer_function2.jpg|thumb|300px|Transfer function showing interpolation artifacts (note the difference in opacity mapping; nearest interpolation is used in both cases)]] | ||
| + | |} | ||
| + | |} | ||
===Module error handling in Slicer=== | ===Module error handling in Slicer=== | ||
Revision as of 14:22, 9 January 2009
Home < 2009 Winter Project Week ChangeTracker Return to Project Week Main Page |
Key Investigators
- Andriy Fedorov, SPL
- Jim Miller, GE
- Karthik Krishnan, Kitware
- Marcel Prastawa, SCI
- Wendy Plesniak, SPL
- Luis Ibanez, Kitware
- Curtis Lisle, Knowledgevis
Objective
ChangeTracker is a software tool for quantification of the subtle changes in pathology. The module provides a workflow pipeline that combines user input with the medical data. As a result we provide quantitative volumetric measurements of growth/shrinkage together with the volume rendering of the tumor and color-coded visualization of the tumor growth/shrinkage.
The objective at this meeting is to determine the status of the Slicer3 components needed for enhanced functionality of ChangeTracker, identify new uses of ChangeTracker, and propose the tentative roadmap for ChangeTracker evolution for the next 6 months.
Approach, Plan
We consider the following improvements to ChangeTracker:
- Extending the interface to allow analysis of more than two images
- Volume rendering of tumor segmentation: visualization and 3d widget for volume cropping
- Using Lightbox for tumor progression visualization
- Robustness of initial tumor segmentation. Currently, we use user-guided thresholding. It must be quick, with minimum input from the user. Discuss possible approaches with Marcel, other groups in tumor segmentation research.
- Consider error handling interface issues in execution of an external module (how to properly handle registration errors?)
Progress
- Tumor segmentation:discussed with Luis and Karthik capabilities of LesionSizingToolkit: potentially very useful for improved accuracy of meningioma segmentation. Currently in the process of evaluating the performance of the feature detection on meningioma data
- Volume rendering: worked with Karthik, Will, Curt and Steve on fine-tuning label data volume rendering (see below for details). Evaluated different approaches for visualizing tumor and growth results.
- CompareView for results visualization: worked with Jim to implement changes to configure the layout with CompareView, but still debugging setting images for slice views
- Module error handling: discussed with Jim changes to the module error handling in Slicer (see below for the proposed modifications)
- Time-series analysis functionality in Slicer: 5-year development roadmap within NAC project. Following discussions with Wendy and Steve, currently there are no specifications or finalized plans. Advantages: improved memory efficiency and data access. Short term solution -- augmenting ChangeTracker with the additional wizard step to specify the number of timepoints.
- Longitudinal lesion analysis in Lupus: put together a requirements document with the desired features in ChangeTracker (or ChangeTracker derivative)
"Customer" projects
Relevant projects of interest
- Longitudinal lesion analysis
- 3d widgets
- Volume rendering
- Command line program testing
- Lesion sizing toolkit
Volume rendering of label data
|
We investigated the behavior of volume rendering on the sample data from tumor growth analysis. We identified that it is crucial to use nearest neighbor interpolation for label volume rendering. It appears that the behavior of the opacity mapping function is rather unexplained (see the screenshots):
Programmatic initialization of the opacity transfer function for two-label data: vtkPiecewiseFunction* opacity = vtkPiecewiseFunction::New();
double* imgRange = this->Render_Image->GetPointData()->GetScalars()->GetRange();
opacity->RemoveAllPoints();
opacity->AddPoint(imgRange[0], 0.0);
opacity->AddPoint(min - .1, 0.0);
opacity->AddPoint(min, 1., .5, 1.);
opacity->AddPoint(min+0.1, 0.);
opacity->AddPoint(max-0.1, 0.);
opacity->AddPoint(max, 1., .5, 1.);
if (max < imgRange[1]) {
opacity->AddPoint(max + .1, 0.);
if (max+.1 < imgRange[1]) {
opacity->AddPoint(max+.1, 0.);
opacity->AddPoint(imgRange[1], 0);
}
}
opacity->ClampingOff();
The datasets for testing are available here, and the transfer function can be seen on the screenshots. Current solutions/workarounds we consider: use surface rendering from Marching cubes instead of volume rendering. |
|
Module error handling in Slicer
Problem: current implementation of Slicer does not handle the case when a module is executed as a shared library, and fails with an exception. Currently, the user will not be notified about the problem that occured during module execution. Also, if a module is executed from within another module, there is no mechanism to catch the exception and try to determine what was the source of the problem.
Proposed solution:
- (1) augment the MRMLCommandLineModuleNode with a bool flag that will instruct the module logic whether the exception should be re-thrown or not (this can be done only in the same-thread (ApplyAndWait()) execution mode)
- (2) add the code in CommandLineModuleLogic to set the execution status to error if an exception occurs
References
- E.Konukoglu, W.M.Wells, S.Novellas, N.Ayache, R.Kikinis, P.M.Black, K.M.Pohl. Monitoring Slowly Evolving Tumors. Proc. of 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2008, pp.812-815 link