Difference between revisions of "2010 CCA Retreat"
From NAMIC Wiki
| (31 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
=Introduction= | =Introduction= | ||
| − | This is a one-day retreat for the clinical computational anatomy project. The purpose is to review the state of this project and formulate strategic plans. | + | This is a one-day retreat for the clinical computational anatomy project of NAC. The purpose is to review the state of this project, learn about related external ontological efforts, and formulate strategic plans for the rest of the funding cycle. |
=Date and Location:= | =Date and Location:= | ||
*June 18 2010 | *June 18 2010 | ||
*1249 Boylston St. 2nd floor demo room | *1249 Boylston St. 2nd floor demo room | ||
| + | *Start at 8.30 | ||
=Agenda= | =Agenda= | ||
| − | * | + | [[image:CCAFeb2010.png|600px|right]] |
| − | ** | + | ==Introduction== |
| − | ** | + | *CCA specific aims: See non-public Feb 2010 [https://intweb.spl.harvard.edu/images/7/76/Talos-NAC-EAB-2010.ppt presentation] |
| − | ** | + | *MRI image-based atlas |
| − | * | + | ** Structural data and rough segmentation on pubdb |
| − | *Ontology tools | + | ** DWI needs to be re-acquired |
| + | ** Anatomical Hierarchy still in development | ||
| + | *linking imaging and ontological data | ||
| + | ** model hierarchy from February no longer being used | ||
| + | ** new label map color table being used to drive the model naming and hierarchy | ||
| + | ** some freesurfer labels are not well defined or organized | ||
| + | ** need a structured way to deal with the clash of concepts from different labeling schemes | ||
| + | ** wiki label lists used to generate slicer color tables | ||
| + | ** color tables are not hierarchical | ||
| + | ** need to match labeling scheme to the application domain (e.g. em segmenter brain segmentation) | ||
| + | ** some form of integer is typically used by researchers to generate labels | ||
| + | ** ideally these would map to unique concepts that are well defined | ||
| + | ** guide for developers rather than having them pick arbitrary numbers | ||
| + | ** we can provide a standard structure an then map unconstrained input data into our structure | ||
| + | ** discussion of the need for UUIDs as an indirection between labelmaps and, for example, radlex IDs | ||
| + | ** how does this related to [https://cabig.nci.nih.gov/tools/AIM AIM] for describing image metadata? | ||
| + | ** general agreement that we need to integrate our efforts in the context of AIM (describe the context of the acquisition and subject) | ||
| + | ** demo of current FMA browser from GE | ||
| + | ** discussion of bioportal and other visualization approaches | ||
| + | ** demo of JSON/sqlite FMA encoding | ||
| + | ** demo of python triple table package | ||
| + | ** comparison with jenna and gui on bioportal | ||
| + | *brain function ontology | ||
| + | ** discussion of Jessica Turner's CCOG project | ||
| + | *data visualization | ||
| + | ** reviewed functionality of query atlas | ||
| + | |||
| + | ==Current Progress== | ||
| + | *2009-10 brain atlas (Ron Kikinis, Michael Halle, Daniel Rubin) | ||
| + | **data acquisition | ||
| + | **labeling | ||
| + | **thalamus labeling | ||
| + | |||
| + | *naming and color assignment | ||
| + | **[http://wiki.slicer.org/slicerWiki/index.php/Slicer3:LUTs_and_Ontologies wiki-based slicer color files] | ||
| + | **CCA brain hierarchy | ||
| + | **new default slicer LUT | ||
| + | |||
| + | *extracting data from ontologies | ||
| + | **JSONTO data format and embedded sqlite database | ||
| + | **FMA, RadLex, NIFstd conversion | ||
| + | **example uses | ||
| + | |||
| + | *Ontology visualization | ||
| + | **java-based ontology visualization tool (Jim Miller) | ||
| + | |||
| + | *Integration with Slicer | ||
| + | **QueryAtlas (Steve Pieper) | ||
| + | |||
| + | ==Discussion== | ||
| + | *Current state of relevant ontologies and tools (Daniel Ruben) | ||
| + | |||
| + | *Making ontologies useable | ||
| + | **How do we factor out complete and useful information from relevant ontologies and present it do developers/researchers/users? | ||
| + | **How can this information be integrated into the Slicer/NA-MIC infrastructure? | ||
| + | ***JSONTO reader | ||
| + | ***VTK infovis | ||
| + | ***Query atlas extensions | ||
| + | ***Segmentation | ||
| + | **What's the best way to encourage researchers to use and create knowledge in machine-readable format? | ||
| + | **(Your questions here!) | ||
| + | |||
| + | ==Final discussion and wrap up== | ||
| + | * Possible Anatomist Collaborators | ||
=Participants= | =Participants= | ||
*Ron Kikinis | *Ron Kikinis | ||
*Michael Halle | *Michael Halle | ||
| − | |||
*Daniel Rubin | *Daniel Rubin | ||
*Jim Miller | *Jim Miller | ||
*Steve Pieper | *Steve Pieper | ||
| − | * | + | *Nicole Aucoin |
| + | *Dominik Meier | ||
Latest revision as of 18:12, 18 June 2010
Home < 2010 CCA RetreatContents
Introduction
This is a one-day retreat for the clinical computational anatomy project of NAC. The purpose is to review the state of this project, learn about related external ontological efforts, and formulate strategic plans for the rest of the funding cycle.
Date and Location:
- June 18 2010
- 1249 Boylston St. 2nd floor demo room
- Start at 8.30
Agenda
Introduction
- CCA specific aims: See non-public Feb 2010 presentation
- MRI image-based atlas
- Structural data and rough segmentation on pubdb
- DWI needs to be re-acquired
- Anatomical Hierarchy still in development
- linking imaging and ontological data
- model hierarchy from February no longer being used
- new label map color table being used to drive the model naming and hierarchy
- some freesurfer labels are not well defined or organized
- need a structured way to deal with the clash of concepts from different labeling schemes
- wiki label lists used to generate slicer color tables
- color tables are not hierarchical
- need to match labeling scheme to the application domain (e.g. em segmenter brain segmentation)
- some form of integer is typically used by researchers to generate labels
- ideally these would map to unique concepts that are well defined
- guide for developers rather than having them pick arbitrary numbers
- we can provide a standard structure an then map unconstrained input data into our structure
- discussion of the need for UUIDs as an indirection between labelmaps and, for example, radlex IDs
- how does this related to AIM for describing image metadata?
- general agreement that we need to integrate our efforts in the context of AIM (describe the context of the acquisition and subject)
- demo of current FMA browser from GE
- discussion of bioportal and other visualization approaches
- demo of JSON/sqlite FMA encoding
- demo of python triple table package
- comparison with jenna and gui on bioportal
- brain function ontology
- discussion of Jessica Turner's CCOG project
- data visualization
- reviewed functionality of query atlas
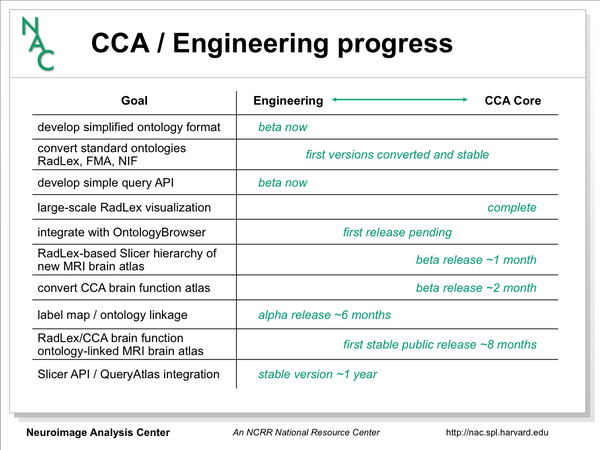
Current Progress
- 2009-10 brain atlas (Ron Kikinis, Michael Halle, Daniel Rubin)
- data acquisition
- labeling
- thalamus labeling
- naming and color assignment
- wiki-based slicer color files
- CCA brain hierarchy
- new default slicer LUT
- extracting data from ontologies
- JSONTO data format and embedded sqlite database
- FMA, RadLex, NIFstd conversion
- example uses
- Ontology visualization
- java-based ontology visualization tool (Jim Miller)
- Integration with Slicer
- QueryAtlas (Steve Pieper)
Discussion
- Current state of relevant ontologies and tools (Daniel Ruben)
- Making ontologies useable
- How do we factor out complete and useful information from relevant ontologies and present it do developers/researchers/users?
- How can this information be integrated into the Slicer/NA-MIC infrastructure?
- JSONTO reader
- VTK infovis
- Query atlas extensions
- Segmentation
- What's the best way to encourage researchers to use and create knowledge in machine-readable format?
- (Your questions here!)
Final discussion and wrap up
- Possible Anatomist Collaborators
Participants
- Ron Kikinis
- Michael Halle
- Daniel Rubin
- Jim Miller
- Steve Pieper
- Nicole Aucoin
- Dominik Meier