2010 Summer Project Week MRSI module and SIVIC interface
Key Investigators
- MIT: Bjoern Menze, Mangpo Phothilimthana, Polina Golland
- UCSF: Olson Beck, Jason Crane (Nelson Lab)
Project
Objective
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases -- such as tumors in brain, breast and prostate -- can be can be associated with characteristic changes in the metabolic level.
The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI -- and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities -- in Slicer.
Approach, Plan
Spectral fitting routines have been implemented, using a HSVD filter for water peak removal and baseline estimation, and a constrained non-linear least squares optimization for the fit of the resonance line models. Current fitting routines require, however, several external software libraries not to be distributed, installed and used easily.
We want to integrate the currently available MRSI quantitation routines with the SIVIC Slicer interface developed at the UCSF.
Progress
We approached the integration from two sides:
- We integrated one of the two MRSI signal processing routines (Fig. 1) into a svk/SIVIC library prototype class.
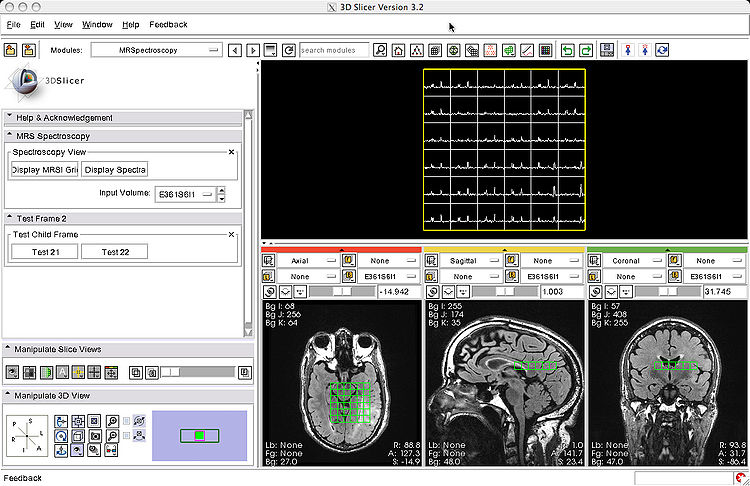
- We integrated the SIVIC visualization routines into Slicer (Fig. 3, screenshot below) as a prototype module.
Further steps will comprise the full integration of the signal processing routines, and the joint display of metabolic maps (Fig. 2) and spectral information within the Slicer module.