2010 Summer Project Week MR to Ultrasound Registration Methodology
Key Investigators
- University Erlangen: Dieter Hahn, Joachim Hornegger
- BWH: Sandy Wells, Tina Kapur, Andriy Fedorov
- Kitware: Stephen Aylward
3D Ultrasound (US) is an emerging diagnostic technique that is non-invasive, fast and comparably cheap. It is commonly used for acquiring images of fetuses or anatomical regions and organs near the skin, for example the thyroid gland or other parenchymatous organs. 3D US has some advantages compared to 2D US, as it the diagnosis may become largely observer-independent and accurate volumetric measurements become possible. Compared to alternative imaging techniques, like CT or MRI, it features a much better spatial resolution and images can be acquired faster and a lot easier. It may, therefore, be combined with another imaging technique in order to enable ongoing screening during treatment or control studies. The combination of MR and US is desirable, as both techniques are very feasible for the analysis of soft tissue. However, this is a challenging task and quite a few problems have to be addressed.
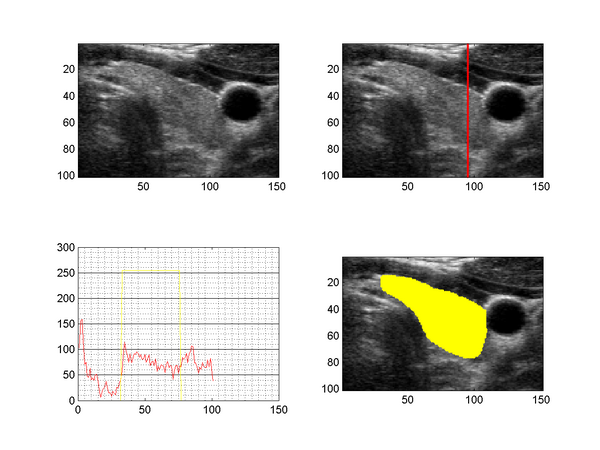
The figure above shows a slice taken from a 3D ultrasound image of the human thyroid (upper left), together with the intensity profile along a line through the tyhroid (upper right, lower left) and a segmentation (lower right). US images suffer from speckle noise, sonic attenuation and also regions of sound shadows (insufficient gel layer). The sonic attenuation cannot be fully compensated by the imaging devices and the effect is clearly visible when examining the intensity profile along a line through the thyroid region (lower left image), which shows decreasing intensities with increasing distance to the surface.
In previous work, we have examined several pre-processing techniques for 3D US denoising, however, we did not find a satisfying solution for the bias field correction of the images. Since the intensity-based registration of MR with 3D US is a very hard problem and only few work has been done on the registration of 3D US with MR images, we will try to come up with some ideas for a combined approach for the bias field correction, segmentation and registration during the project weak.
Objectives
- denoising and bias field correction of 3D US data
- model for the combined image correction, segmentation and registration between 3D US and MRI
- visualization of all processing steps within Slicer
Approach, Plan
The bias field on the 3D US data varies spatially with a very low frequency. It is similar to the bias field effects in MR caused by magnetic inhomogeneities. We will investigate whether an EM-based approach may be used for the correction, similar to the previous work by Wells et. al. and Pohl et. al., and try to adapt the already existing Slicer code to this problem.
For the registration, we may have to come up with a new formulation that uses not only the statistics of the intensities, but also some additional information about the tissue boundaries. The tissue boundaries are directly influencing the visible content of the 3D US images, however, they have a different appearance and structure within the MRI.
Progress
During this week, we have investigated whether the EMSegment module in Slicer can be used for the segmentation of the images. We have made several experiments using phantom data and also real patient images. The focus was mainly on using EMSegment in order to see how an intensity-based EM segmentation looks like on these types of images. Prior to the application of EMSegment, a bias field correction and denoising were performed using Slicer (N4ITK Bias Field Correction, Curvature Anisotropic Diffusion).
The following images show one of the phantom data that we have acquired and the pre-processing results:
From left: slice of 3D US phantom with visible attenuation field, estimated bias field, corrected image, image after denoising. Each image shows only one slice of the corresponding volume.
From our database, we have taken 5 thyroid images for the experiment, which all show a distinct vessel near the thyroid gland, and applied the pre-processing as shown above. For these images, a manual segmentation of the thyroid gland is known. The model has been extended by a segmentation of the vessel, which was achieved using a combination of the modules FastMarchingSegmentation and Robust Statistics Segmentation.
Based on these segmentations, we have created atlases for the prior probabilities of the thyroid and the vessel. The atlases were generated based on Gaussian smoothing, i.e. the labels were smoothed, affinely registered and combined to sum-up to one. For simplicity, the labels have been registered to the first US image, such that the atlas was in the same coordinate space. Five tissue classes were specified: background, thyroid, vessel, muscle and high gain (strong boundaries and fat). We performed a histogram matching of the input images, such that the same statistics of the tissue classes can be assumed throughout the data sets.
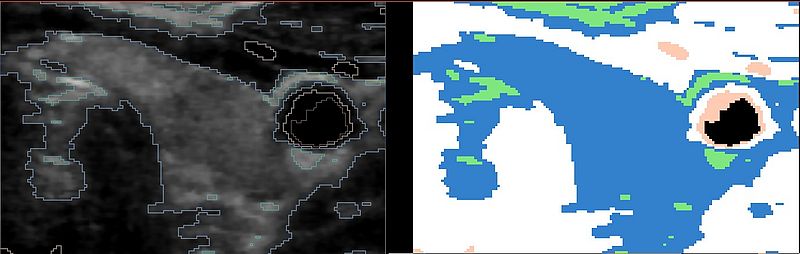
These are the results of the EMSegmenter with just the tissue class distributions:
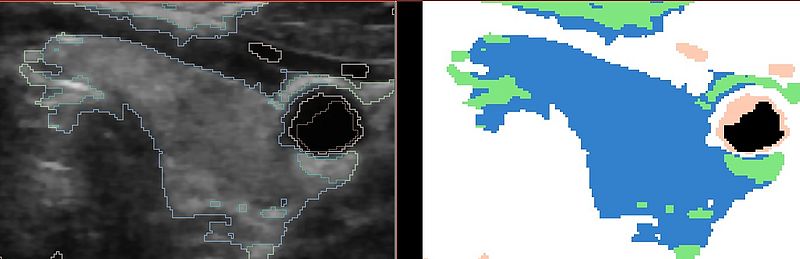
The following result shows the improvement by using the atlas prior on the thyroid gland. The shape is much more constrained than in the previous result:
In future work, we want to further refine the segmentation by generating a shape model for the thyroid and potentially also for the attached vessels. We want to use this segmentation information in order to derive a mathematical formulation for the registration between MR and US. We have some ideas for the mathematical extension of the EM framework, which require further investigation, additional data (MR) and experiments.
Delivery Mechanism
The final work will be provided as ITK modules and it intended to integrate them into a Slicer module.
References
- W.M. Wells, R. Kikinis, W.E.L. Grimson, F. Jolesz. Adaptive segmentation of MRI data. IEEE Transactions on Medical Imaging, 15, pp. 429-442, 1996
- K.M. Pohl, J. Fisher, W.E.L. Grimson, R. Kikinis, and W.M. Wells. A Bayesian model for joint segmentation and registration. NeuroImage, 31(1), pp. 228-239, 2006
- E.N.K. Kollorz, D.A. Hahn, R. Linke, T.W. Goecke, J. Hornegger, and T. Kuwert. Quantification of Thyroid Volume using 3-D Ultrasound Imaging. IEEE Transactions on Medical Imaging, 27(4), pp. 457-466, 2008
- W. Wein, S. Brunke, A. Khamene, M.R. Callstrom, N. Navab. Automatic CT-Ultrasound Registration for Diagnostic Imaging and Image-guided Intervention. Medical Image Analysis, 12(5), pp. 577-585, 2008