Difference between revisions of "2010 Winter Project Week XND"
From NAMIC Wiki
RandyGollub (talk | contribs) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
| + | {| | ||
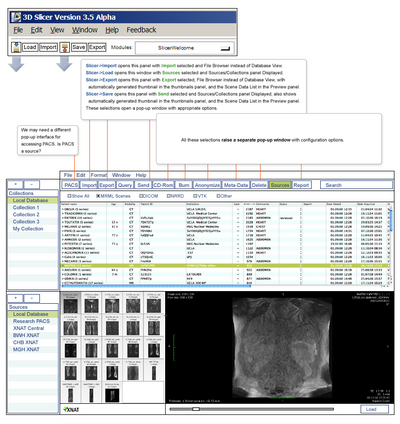
| + | |[[Image:SlicerXNDLayout_Draft2.png|thumb|400px| Upload pipeline of tagged data to remote host]] | ||
| + | |} | ||
| + | |||
==Key Investigators== | ==Key Investigators== | ||
* Wendy Plesniak | * Wendy Plesniak | ||
| Line 8: | Line 13: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | Modify the XNAT Desktop user interface to have a more PACS-like interface while accounting for complexities of research data that aren't part of typical patient-centric interfaces. | + | * Modify the XNAT Desktop user interface to have a more PACS-like interface while accounting for complexities of research data that aren't part of typical patient-centric interfaces. |
| + | * Slicer will also have a native PACS-like '''browse/load/save''' interface that will act as a client to XNAT's File Server and other database back ends; we want to ensure as parallel a user experience as possible between Slicer's native interface and XND to help users switch easily between the two. | ||
</div> | </div> | ||
| Line 15: | Line 21: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Use well-received interfaces (Osirix, GE) to motivate the new XND interface. Continue developing mock-ups and iteratively implementing in XND. | + | * Use well-received interfaces (Osirix, GE) to motivate the new XND interface. Continue developing mock-ups and iteratively implementing in XND. |
| + | * Design for multiple levels of expertise (novice, intermediate, expert), and '''focus on needs of (mi2b2) clinician scientist users''' as a key user group | ||
</div> | </div> | ||
| Line 21: | Line 28: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | Wendy developed an initial mock-up that received additional tuning during her recent visit to STL. | + | Wendy developed an initial mock-up that received additional tuning during her recent visit to STL. Subsequent discussions with Ron have revealed additional requirements and design constraints. Work on transcribing this information and storyboard the browse/load DICOM workflow will continue this week. |
| + | |||
| + | Discussions this week have refined the design approach and next steps. | ||
| + | *Will begin with DICOM data, but will accommodate other data types that slicer supports. | ||
| + | *Will provide three data "views": Project, Study, and Subject views. Will get rid of "File view" in XND. | ||
| + | *Wendy will design a set of filters to refine search (worked with Tim and Dave to understand the technical constraints, and learn about ways to generate queries.) | ||
| + | *Slicer side will develop thumbnails for all MRML scenes | ||
| + | *XNAT team will develop thumbnails for DICOM. | ||
| + | *Wendy will design a set of workflows for importing and marking up data (DICOM and other). | ||
| + | *Tim and Dan will discuss MIME type variable | ||
| + | *Tim and Dan will discuss where we assign "MRML" as variable value (Format, Content, Mimetype?) | ||
| + | *Tim and Wendy discussed wiki page for "recommended" variable values for these variables, to promote data sharing. Mechanism for updating metadata if a recommended variable value changes? | ||
| + | *Also discussed designing a "QTWidget" for transacting with XNAT -- useful for many groups. | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 16:49, 8 January 2010
Home < 2010 Winter Project Week XNDKey Investigators
- Wendy Plesniak
- Ron Kikinis
- Dan Marcus
Objective
- Modify the XNAT Desktop user interface to have a more PACS-like interface while accounting for complexities of research data that aren't part of typical patient-centric interfaces.
- Slicer will also have a native PACS-like browse/load/save interface that will act as a client to XNAT's File Server and other database back ends; we want to ensure as parallel a user experience as possible between Slicer's native interface and XND to help users switch easily between the two.
Approach, Plan
- Use well-received interfaces (Osirix, GE) to motivate the new XND interface. Continue developing mock-ups and iteratively implementing in XND.
- Design for multiple levels of expertise (novice, intermediate, expert), and focus on needs of (mi2b2) clinician scientist users as a key user group
Progress
Wendy developed an initial mock-up that received additional tuning during her recent visit to STL. Subsequent discussions with Ron have revealed additional requirements and design constraints. Work on transcribing this information and storyboard the browse/load DICOM workflow will continue this week.
Discussions this week have refined the design approach and next steps.
- Will begin with DICOM data, but will accommodate other data types that slicer supports.
- Will provide three data "views": Project, Study, and Subject views. Will get rid of "File view" in XND.
- Wendy will design a set of filters to refine search (worked with Tim and Dave to understand the technical constraints, and learn about ways to generate queries.)
- Slicer side will develop thumbnails for all MRML scenes
- XNAT team will develop thumbnails for DICOM.
- Wendy will design a set of workflows for importing and marking up data (DICOM and other).
- Tim and Dan will discuss MIME type variable
- Tim and Dan will discuss where we assign "MRML" as variable value (Format, Content, Mimetype?)
- Tim and Wendy discussed wiki page for "recommended" variable values for these variables, to promote data sharing. Mechanism for updating metadata if a recommended variable value changes?
- Also discussed designing a "QTWidget" for transacting with XNAT -- useful for many groups.