Difference between revisions of "2011 Summer Project Week Customizing EMSegmenter pipelines for brain lesions"
From NAMIC Wiki
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| Line 45: | Line 45: | ||
==References== | ==References== | ||
| + | |||
| + | <gallery perrow=1: widths=1250px : heights=400px> | ||
| + | Image:EMS LesionExp T1.png | ||
| + | Image:EMS LesionExp FLAIR.png | ||
| + | Image:EMS LesionExp Segmentation.png | ||
| + | </gallery> | ||
| + | |||
http://www.slicer.org/slicerWiki/index.php/EMSegmenter-Tasks <br> | http://www.slicer.org/slicerWiki/index.php/EMSegmenter-Tasks <br> | ||
| + | |||
<!-- Add links here to background material: where can we download the data? where is the source code repository? what papers or web sites will help someone quickly learn what the outstanding issues are? --> | <!-- Add links here to background material: where can we download the data? where is the source code repository? what papers or web sites will help someone quickly learn what the outstanding issues are? --> | ||
Revision as of 16:55, 20 June 2011
Home < 2011 Summer Project Week Customizing EMSegmenter pipelines for brain lesions- Notfound.png
Interesting picture to be added...
Customizing EMSegmenter pipelines for brain lesions
Key Investigators
- University of Pennsylvania: Dominique Belhachemi
- University of Pennsylvania: Kilian Pohl
- Brigham and Women's Hospital: Alex Zaitsev
Objective
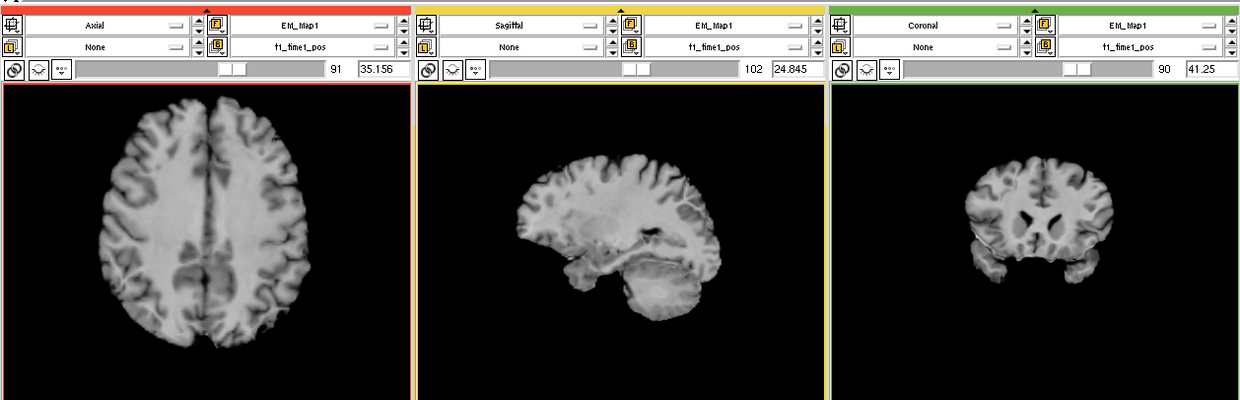
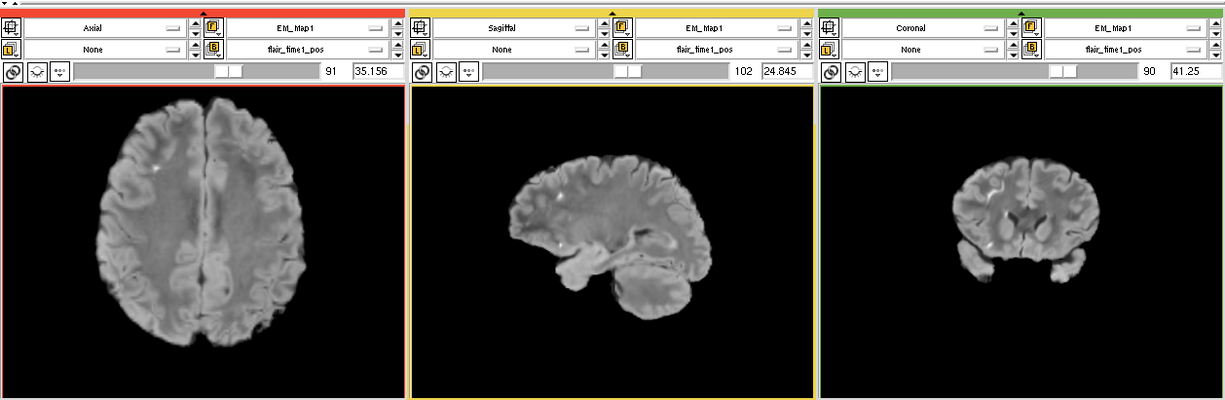
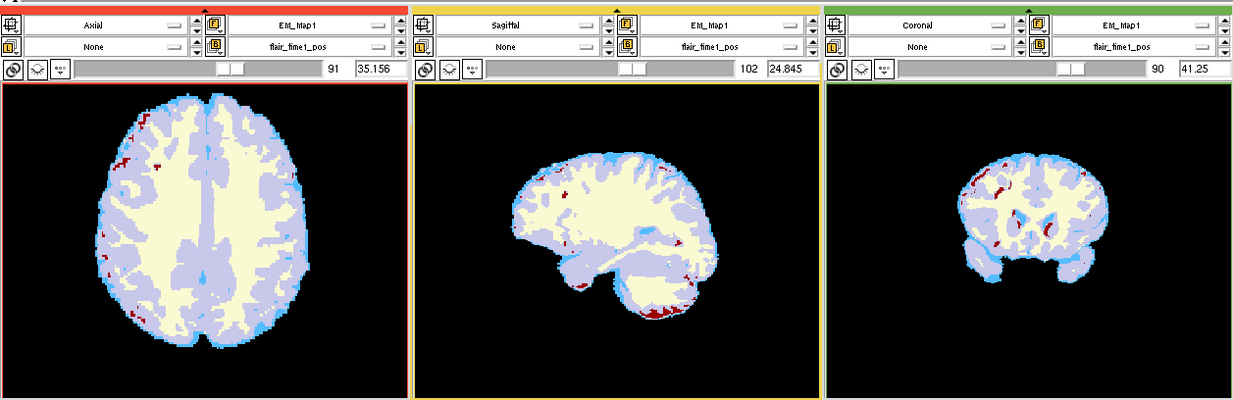
The goal is to segment white matter lesions based on T1 and FLAIR brain scans.
Approach, Plan
We want to use the EMSegmenter modle for this project. The plan is to create a two channel pipeline which includes pre-processing (e.g. registration) and applying the EM algorithm to perform the segmentation.
Progress
References
http://www.slicer.org/slicerWiki/index.php/EMSegmenter-Tasks
Delivery Mechanism
This work will be delivered to the NAMIC Kit as a
- Slicer Module
- Built-in:
- Extension -- commandline:
- Extension -- loadable: