Difference between revisions of "2011 Summer Project Week DICOM RT"
| (38 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | ||
| − | |||
</gallery> | </gallery> | ||
| + | |||
| + | [[Image:sshot.png|300px|Synthetic noise]] | ||
| + | |||
| + | Synthetic DICOM-RT sample data [http://forge.abcd.harvard.edu/gf/download/frsrelease/85/1865/random.zip] | ||
==Key Investigators== | ==Key Investigators== | ||
| Line 9: | Line 12: | ||
* Kitware: Luis Ibanez | * Kitware: Luis Ibanez | ||
* Isomics: Steve Pieper | * Isomics: Steve Pieper | ||
| + | * Cosmo software: Drew Wassem | ||
| + | * A*STAR: Alexandre GOUAILLARD (in spirit) | ||
| + | |||
| + | '''Synthetic images, vector fields, RT structures and RT doses in Slicer and ITK''' | ||
Figure out the best representation in ITK and Slicer4 for various RT and related datastructures. | Figure out the best representation in ITK and Slicer4 for various RT and related datastructures. | ||
| Line 15: | Line 22: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
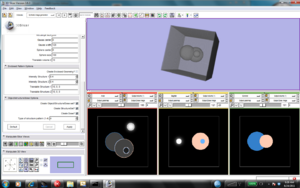
| − | + | We are designing synthetic DICOM-RT dataset which includes CT image, structure set and dose distribution. The following guidelines are used. We measured the noise power spectrum of a real CT image and constructed synthetic image with exactly the same spectrum. Then we included "skin" and one internal spherical structure. Synthetic contours were generated and recorded in labelmap format. Synthetic dose distribution was generated in mha format. | |
| − | |||
| Line 24: | Line 30: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | Currently synthetic DICOM-RT is loaded into Slicer 3.6.3 from file through Plastimatch extension DICOM/DICOM-RT Import. We plan to build Plastimatch command line module that will allow us to create synthetic image, structures and dose distribution interactively according to user's needs. | |
| − | |||
| − | |||
</div> | </div> | ||
| Line 33: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Discussed the use of Slicer4 hierarchies to group structure sets | |
| + | * Reviewed DICOM networking interface with respect to RT workflows | ||
| + | * Worked around resampling performance issues with itk vector images | ||
Latest revision as of 14:02, 24 June 2011
Home < 2011 Summer Project Week DICOM RTSynthetic DICOM-RT sample data [1]
Key Investigators
- MGH: Nadya Shusharina, Greg Sharp
- Kitware: Luis Ibanez
- Isomics: Steve Pieper
- Cosmo software: Drew Wassem
- A*STAR: Alexandre GOUAILLARD (in spirit)
Synthetic images, vector fields, RT structures and RT doses in Slicer and ITK
Figure out the best representation in ITK and Slicer4 for various RT and related datastructures.
Objective
We are designing synthetic DICOM-RT dataset which includes CT image, structure set and dose distribution. The following guidelines are used. We measured the noise power spectrum of a real CT image and constructed synthetic image with exactly the same spectrum. Then we included "skin" and one internal spherical structure. Synthetic contours were generated and recorded in labelmap format. Synthetic dose distribution was generated in mha format.
Approach, Plan
Currently synthetic DICOM-RT is loaded into Slicer 3.6.3 from file through Plastimatch extension DICOM/DICOM-RT Import. We plan to build Plastimatch command line module that will allow us to create synthetic image, structures and dose distribution interactively according to user's needs.
Progress
- Discussed the use of Slicer4 hierarchies to group structure sets
- Reviewed DICOM networking interface with respect to RT workflows
- Worked around resampling performance issues with itk vector images
References
Delivery Mechanism
This work will be delivered to the NAMIC Kit as a
- NITRIC distribution
- Slicer Module
- Built-in: perhaps RT support can become part of MRML
- Extension -- commandline: Maybe, if needed
- Extension -- loadable: yes, for a custom interface