Difference between revisions of "2011 Summer Project Week NerveSeg"
| Line 6: | Line 6: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2011.png|[[2011_Summer_Project_Week#Projects|Projects List]] | ||
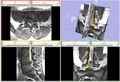
| + | Image:NergeSeg-scr1.png|Figure 1 - Example of Manual Nerve Segmentation | ||
</gallery> | </gallery> | ||
<gallery> | <gallery> | ||
| − | |||
| − | |||
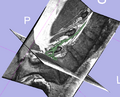
Image:NerveSeg-scr3.png|Figure 2 - Example_1: non-cleaned results. | Image:NerveSeg-scr3.png|Figure 2 - Example_1: non-cleaned results. | ||
Revision as of 21:18, 23 June 2011
Home < 2011 Summer Project Week NerveSegNerve Segmentation
- Successful segmentation including afterall
Key Investigators
- MIT: Adrian Dalca, Polina Golland
- BWH: Giovanna Danagoulian, Ehud Schmidt
- SPL: Ron Kikinis
Objective
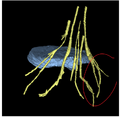
We are developing a nerve segmentation algorithm for the automatic isolation of nerves and nerve ganglia inside the spinal sack and out through the vertebrae in new MR Myelography images. Current progress can track the core of a Nerve.
Approach, Plan
Currently we use a particle-filter tracking approach for segmenting the nerves. The algorithm is given a seed point, preferably somewhere in the spine. The particles are tubes following Bézier curves (and hence forming a B-spline track). The dynamics model encourages continuity and smoothness. The image likelihood model compares gradient fields and intensities of predicted patches with image observations to evaluate a posterior distribution of the particles' importance. While we can currently usually track the nerve cores, usually fully throughout the vertebral canal, the algorithm does not delineate the full extent of the nerves, and behaves poorly on peripheral nerves outside of the vertebral canal. We will modify the likelihood function and parameters and concentrate on achieving this during the project week.
Progress