Difference between revisions of "2012 Summer Project Week:Intraoperative Tract Detection"
| (10 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | ||
| − | Image:PreliminaryWorkFiberSimilarity. | + | Image:PreliminaryWorkFiberSimilarity.png|Fiber tract comparison to define similarity. |
| − | Image: | + | Image:BrainShiftDiagram.png|Challenge of brain shift |
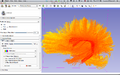
| + | Image:Tracts_colored_by_distance_to_arcuate.png|Slicer-Python result during project week | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * BWH: Lauren O'Donnell, Isaiah Norton, Alex Golby | + | * BWH: Lauren O'Donnell, Isaiah Norton, Alex Golby, Steve Pieper |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 22: | Line 23: | ||
Our approach for tract detection is to compare new tracts to the prior information (preoperative plan). | Our approach for tract detection is to compare new tracts to the prior information (preoperative plan). | ||
| − | Our plan for the project week is to integrate Python code into a slicer module | + | Our plan for the project week is to integrate our Python code that has been developed for this project into a slicer module. |
| + | This will enable clinical testing and validation of the methods. | ||
</div> | </div> | ||
| Line 29: | Line 31: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | We have successfully imported our python package into Slicer. (Some aspects of its dependencies are unresolved, the optimization routines in scipy that we use for tract-based registration are not currently available in Slicer and we have met with Steve Pieper about using system Python and compiled his version of Slicer that may enable this moving forward). A new intial Slicer module GUI now allows comparison of whole-brain tracts to a tract of interest, see result image above. | |
| − | |||
</div> | </div> | ||
| Line 39: | Line 40: | ||
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below) | This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below) | ||
| − | |||
#Slicer Module | #Slicer Module | ||
##Built-in | ##Built-in | ||
| Line 47: | Line 47: | ||
==References== | ==References== | ||
| − | * | + | * O'Donnell LJ, Wells WM, Golby A, Westin CF. Unbiased groupwise registration of white matter tractography. To appear in Medical Image Computing and Computer Assisted Intervention. 2012. |
Latest revision as of 13:56, 22 June 2012
Home < 2012 Summer Project Week:Intraoperative Tract DetectionKey Investigators
- BWH: Lauren O'Donnell, Isaiah Norton, Alex Golby, Steve Pieper
Objective
We are developing methods for intraoperative tract detection based on a preoperative plan.
Approach, Plan
Our approach for tract detection is to compare new tracts to the prior information (preoperative plan). Our plan for the project week is to integrate our Python code that has been developed for this project into a slicer module. This will enable clinical testing and validation of the methods.
Progress
We have successfully imported our python package into Slicer. (Some aspects of its dependencies are unresolved, the optimization routines in scipy that we use for tract-based registration are not currently available in Slicer and we have met with Steve Pieper about using system Python and compiled his version of Slicer that may enable this moving forward). A new intial Slicer module GUI now allows comparison of whole-brain tracts to a tract of interest, see result image above.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- Slicer Module
- Built-in
- Extension -- commandline
- Extension -- loadable
- Other (Please specify)
References
- O'Donnell LJ, Wells WM, Golby A, Westin CF. Unbiased groupwise registration of white matter tractography. To appear in Medical Image Computing and Computer Assisted Intervention. 2012.