Difference between revisions of "2012 Summer Project Week:XTK"
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2012.png|Projects List Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus…') |
|||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | ||
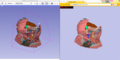
| − | Image: | + | Image:Xtkabdom.png|The SPL Abdominal Atlas. ''Left'': In 3D Slicer version 4, ''Right'': In a web browser exported using the '''WebGL Export''' module. |
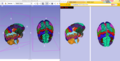
| − | Image: | + | Image:Xtkbrain.png|The NAC Brain Atlas: ''Left'': In Slicer, ''Right'': In a web browser. |
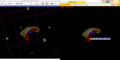
| + | Image:Xtkblack.png|An example WebGL export with black background color and a caption tooltip. ''Left'': In Slicer, ''Right'': In a web browser. | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * Boston Children's Hospital: Daniel Haehn, Nicolas Rannous, Rudolph Pienaar |
| − | * | + | * Isomics Inc.: Steve Pieper |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 15: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | XTK is a WebGL framework providing an easy-to-use API to visualize scientific data on the web. No background or knowledge in Computer Graphics is required and a [https://github.com/xtk/X/wiki/X:Fileformats variety of file formats] can be natively read and visualized with very little JavaScript code.<br><br>Nevertheless, some researchers want to share their data without writing any code so we propose a '''one-click WebGL export module''' for 3D Slicer 4. | |
| − | |||
| − | |||
| − | |||
| Line 33: | Line 24: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | The '''WebGL Export''' module should enable the export of ''Surface Models'', ''Fiber Tracks'' and ''Volumes in 2D and 3D (including Volume Rendering)'' from 3D Slicer 4 to the web. | ||
| − | + | First step of further development is to update the currently existing '''WebGL Export''' module to a new XTK version. Additionally, a TrackVis file format (.TRK) writer should be implemented for 3D Slicer 4 since XTK natively supports this format for Tractography. | |
| − | + | Also, it should be interesting to investigate if Slicer Data Bundles or Scene Views can be exported. | |
</div> | </div> | ||
| Line 43: | Line 35: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | A first version of the '''WebGL Export''' module is part of the latest 3D Slicer 4 release. Currently, it supports the export of ''Surface Models'' to a website (see [http://www.na-mic.org/Wiki/index.php/File:WebGLExport_TutorialContestSummer2012.pdf tutorial]). | |
| − | |||
</div> | </div> | ||
| Line 51: | Line 42: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | This work will be delivered to the NA-MIC Kit as a | + | This work will be delivered to the NA-MIC Kit as a |
#ITK Module | #ITK Module | ||
| Line 57: | Line 48: | ||
##Built-in | ##Built-in | ||
##Extension -- commandline | ##Extension -- commandline | ||
| − | ##Extension -- loadable | + | ##Extension -- loadable '''YES''' |
#Other (Please specify) | #Other (Please specify) | ||
==References== | ==References== | ||
| − | * | + | * [http://goXTK.com The XTK website] including lessons, demos etc. |
| − | * | + | * [https://github.com/xtk/SlicerWebGLExport Github Project Page] for the 3D Slicer WebGL Export module |
| − | + | * '''WebGL for Baby Brains: Neuroimaging in the Browser''' at the WebGL Camp Orlando 2012, [http://www.youtube.com/watch?v=SQuH-xpdlbc Video] | |
| − | * | + | * '''Science!''' featuring XTK, [http://creativejs.com/2012/04/science/ CreativeJS.com] |
| + | * '''The X Toolkit for Scientific Visualization''' on [http://www.webgl.com/2012/03/webgl-demo-the-x-toolkit-for-scientific-visualization/ WebGL.com] | ||
| + | * D. Haehn, N. Rannou, B. Ahtam, E. Grant, R. Pienaar: '''Neuroimaging in the Browser using the X Toolkit''', INCF Neuroinformatics 2012, [http://www.neuroinformatics2012.org/abstracts/neuroimaging-in-the-browser-using-the-x-toolkit Abstract] | ||
| + | * A. Klein, N. Nichols, D. Haehn: '''Mindboggle 2 interface: online visualization of extracted brain features with XTK''', INCF Neuroinformatics 2012, [http://www.neuroinformatics2012.org/abstracts/mindboggle-2-interface-online-visualization-of-extracted-brain-features-with-xtk Abstract] | ||
Revision as of 20:48, 15 June 2012
Home < 2012 Summer Project Week:XTKKey Investigators
- Boston Children's Hospital: Daniel Haehn, Nicolas Rannous, Rudolph Pienaar
- Isomics Inc.: Steve Pieper
Objective
XTK is a WebGL framework providing an easy-to-use API to visualize scientific data on the web. No background or knowledge in Computer Graphics is required and a variety of file formats can be natively read and visualized with very little JavaScript code.
Nevertheless, some researchers want to share their data without writing any code so we propose a one-click WebGL export module for 3D Slicer 4.
Approach, Plan
The WebGL Export module should enable the export of Surface Models, Fiber Tracks and Volumes in 2D and 3D (including Volume Rendering) from 3D Slicer 4 to the web.
First step of further development is to update the currently existing WebGL Export module to a new XTK version. Additionally, a TrackVis file format (.TRK) writer should be implemented for 3D Slicer 4 since XTK natively supports this format for Tractography.
Also, it should be interesting to investigate if Slicer Data Bundles or Scene Views can be exported.
Progress
A first version of the WebGL Export module is part of the latest 3D Slicer 4 release. Currently, it supports the export of Surface Models to a website (see tutorial).
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a
- ITK Module
- Slicer Module
- Built-in
- Extension -- commandline
- Extension -- loadable YES
- Other (Please specify)
References
- The XTK website including lessons, demos etc.
- Github Project Page for the 3D Slicer WebGL Export module
- WebGL for Baby Brains: Neuroimaging in the Browser at the WebGL Camp Orlando 2012, Video
- Science! featuring XTK, CreativeJS.com
- The X Toolkit for Scientific Visualization on WebGL.com
- D. Haehn, N. Rannou, B. Ahtam, E. Grant, R. Pienaar: Neuroimaging in the Browser using the X Toolkit, INCF Neuroinformatics 2012, Abstract
- A. Klein, N. Nichols, D. Haehn: Mindboggle 2 interface: online visualization of extracted brain features with XTK, INCF Neuroinformatics 2012, Abstract