Difference between revisions of "2012 Winter Project Week:PelvicRegistration"
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | Project Title: '''Surface-based 3D deformable registration of ultrasound and MRI prostate volumes''' | |
<gallery> | <gallery> | ||
Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | ||
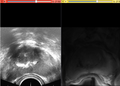
| + | File:Mr us.JPG| | ||
</gallery> | </gallery> | ||
| + | |||
| + | |||
==Investigators at Brigham and Women's == | ==Investigators at Brigham and Women's == | ||
| Line 19: | Line 22: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | We | + | |
| − | penalty for the deformation. Ultrasound images are collected using a stepper, with fixed slice spacing in the axial view. The object is segmented (contoured) in both MRI and ultrasound images and a surface model is created. | + | We will first examine solutions available in Slicer. |
| + | |||
| + | We have also implemented a surface based registration algorithm with elastic energy as the penalty for the deformation. Ultrasound images are collected using a stepper, with fixed slice spacing in the axial view. The object is segmented (contoured) in both MRI and ultrasound images and a surface model is created. | ||
| + | |||
| + | |||
| + | |||
| Line 29: | Line 37: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | During the project week, two solutions were applied to an available dataset. The dataset included three sets of 3D ultrasound and MRI images from three different cases. For all these cases manual segmentation provided by a clinician was available. 1) We used Slicer 4 to implement image-based 3D deformable registration of image labels (results of segmentation) and then apply the resulting warping to the moving (MRI) image. 2) We used the Slicer 3.6 implementation of ICP registration to align the 3D models created from the segmented labels from MRI and ultrasound. Based on this initial alignment, we equalized the length of the base-apex axis of the prostate gland in the two modalities. Then, we updated the label volumes and applied image-based registration (Rigid, affine and deformable) to the updated label maps. | |
| + | Validation is a challenge as landmarks that can be matched from ultrasound and MRI are hard to find in the prostate gland. In one case, calcification is visible in both modalities. Based solely on this landmark, solution (2) appears more accurate. We are planning to consult with clinicians and acquire their feedback on the comparisons of these methods, and validation methods. | ||
| + | <gallery> | ||
| + | File:Mr_us_pros.PNG|[[MR to ultrasound registration, using solutions (1) described in the (progress) section above.]] | ||
| + | </gallery> | ||
</div> | </div> | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
Latest revision as of 07:56, 13 January 2012
Home < 2012 Winter Project Week:PelvicRegistrationProject Title: Surface-based 3D deformable registration of ultrasound and MRI prostate volumes
Investigators at Brigham and Women's
- Computational core: Mehdi Moradi, Firdaus Janoos, Tina Kapur, Jan Egger, Sandy Wells
- Clinical core: Clare Tempany, Paul Nguyen
- Prostate core: Andriy Fedorov
- Present team members in SLC: Mehdi Moradi, Andriy Fedorov, Jan Egger.
Objective
We will implement a basic surface-based deformable registration solution to register prostate 3D ultrasound data to MRI. This is to enable clinical integration of the diagnostic MRI data with ultrasound during prostate brachytherapy with dynamic intraoperative dose planning.
Approach, Plan
We will first examine solutions available in Slicer.
We have also implemented a surface based registration algorithm with elastic energy as the penalty for the deformation. Ultrasound images are collected using a stepper, with fixed slice spacing in the axial view. The object is segmented (contoured) in both MRI and ultrasound images and a surface model is created.
Progress
During the project week, two solutions were applied to an available dataset. The dataset included three sets of 3D ultrasound and MRI images from three different cases. For all these cases manual segmentation provided by a clinician was available. 1) We used Slicer 4 to implement image-based 3D deformable registration of image labels (results of segmentation) and then apply the resulting warping to the moving (MRI) image. 2) We used the Slicer 3.6 implementation of ICP registration to align the 3D models created from the segmented labels from MRI and ultrasound. Based on this initial alignment, we equalized the length of the base-apex axis of the prostate gland in the two modalities. Then, we updated the label volumes and applied image-based registration (Rigid, affine and deformable) to the updated label maps.
Validation is a challenge as landmarks that can be matched from ultrasound and MRI are hard to find in the prostate gland. In one case, calcification is visible in both modalities. Based solely on this landmark, solution (2) appears more accurate. We are planning to consult with clinicians and acquire their feedback on the comparisons of these methods, and validation methods.