Difference between revisions of "2012 Winter Project Week:ScriptView"
From NAMIC Wiki
(Created page with '__NOTOC__ <gallery> Image:PW-SLC2012.png|Projects List Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus…') |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | Image:PW-SLC2012.png|[[2012_Winter_Project_Week#Projects|Projects List]] | ||
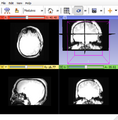
| − | Image: | + | Image:Subj1_II.png|Subject1. |
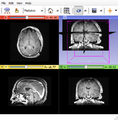
| − | Image: | + | Image:Subj2_II.png|Subject2. |
| + | Image:Subj3_II.png|Subject3. | ||
</gallery> | </gallery> | ||
| − | == | + | ==Key Investigators== |
| − | + | * GE Research: Albert Montillo (montillo@ge.com) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * Isomics: Steve Pieper | |
| − | * | ||
| − | |||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 21: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Programmatically setup 3D Slicer to view data from different subjects while applying the same consistent view. Save screen image to PNG. | |
| − | |||
| − | |||
| − | |||
| Line 34: | Line 26: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Our approach for | + | Our approach is to create a template MRML scene file and reuse it for each subject. MRML scene stores relative paths to items in the scene such as volumes. Python script is passed to Slicer to load the scene per subject and save image.<foo>. |
| − | |||
</div> | </div> | ||
| Line 43: | Line 34: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | The python script has been tested on folder with multiple subject subfolders. All subject folders use same file names for scene elements so that same MRML file works for all subjects. MRML scene need only be setup once, for one subject, then reused for each subject. | |
| − | |||
</div> | </div> | ||
| Line 53: | Line 43: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | This work will be delivered | + | This work will be delivered as a python script. |
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
Latest revision as of 17:02, 13 January 2012
Home < 2012 Winter Project Week:ScriptViewKey Investigators
- GE Research: Albert Montillo (montillo@ge.com)
- Isomics: Steve Pieper
Objective
Programmatically setup 3D Slicer to view data from different subjects while applying the same consistent view. Save screen image to PNG.
Approach, Plan
Our approach is to create a template MRML scene file and reuse it for each subject. MRML scene stores relative paths to items in the scene such as volumes. Python script is passed to Slicer to load the scene per subject and save image.<foo>.
Progress
The python script has been tested on folder with multiple subject subfolders. All subject folders use same file names for scene elements so that same MRML file works for all subjects. MRML scene need only be setup once, for one subject, then reused for each subject.
Delivery Mechanism
This work will be delivered as a python script.