Difference between revisions of "2013 Project Week:WebbasedAnatomicalTeachingFrameworkSummer2013"
From NAMIC Wiki
| Line 18: | Line 18: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
* Over the last several years, an [[CTSC:ARRA_supplement | infrastructure has been developed to integrate health records and PACS image archives]] to support research on large collections of data collected during delivery of healthcare (as contrasted with data collected as part of clinical trials). This resulted in the availability of [http://www.mi2b2.org the mi2b2 system]. An application enabled by this technology is supported by an [http://projectreporter.nih.gov/project_info_description.cfm?aid=8272742&icde=13552329&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC R01 to use clinical scans of children aged zero to six years old to build a teaching atlas]. | * Over the last several years, an [[CTSC:ARRA_supplement | infrastructure has been developed to integrate health records and PACS image archives]] to support research on large collections of data collected during delivery of healthcare (as contrasted with data collected as part of clinical trials). This resulted in the availability of [http://www.mi2b2.org the mi2b2 system]. An application enabled by this technology is supported by an [http://projectreporter.nih.gov/project_info_description.cfm?aid=8272742&icde=13552329&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC R01 to use clinical scans of children aged zero to six years old to build a teaching atlas]. | ||
| − | * One goal of the [http://www.mi2b2.org mi2b2] collaborative effort is to create a web-based teaching tool for fetal and neonatal anatomy as well as a browser for quantified information. A basic prototype has been created: [http://fnndsc.github.io/babybrain/]. The purpose of this project is to construct a 4D MRI dataset as a concatenation of multiple 3D MRI datasets (pediatric MRI scans at different stages of development), and to develop the functionality within | + | * One goal of the [http://www.mi2b2.org mi2b2] collaborative effort is to create a web-based teaching tool for fetal and neonatal anatomy as well as a browser for quantified information. A basic prototype has been created: [http://fnndsc.github.io/babybrain/ http://fnndsc.github.io/babybrain/]. The purpose of this project is to construct a 4D MRI dataset as a concatenation of multiple 3D MRI datasets (pediatric MRI scans at different stages of development), and to develop the functionality within [https://github.com/xtk/X#readme XTK] to visualize and navigate this 4D dataset in a web browser. |
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
Revision as of 18:58, 11 June 2013
Home < 2013 Project Week:WebbasedAnatomicalTeachingFrameworkSummer2013Key Investigators
- Daniel Haehn, Rudolph Pienaar, Ellen Grant (Boston Children's Hospital)
- Steve Pieper (Isomics Inc.)
- Randy Gollub, Lilla Zollei, Nathaniel Reynolds (MGH)
Project Description
Objective
- Over the last several years, an infrastructure has been developed to integrate health records and PACS image archives to support research on large collections of data collected during delivery of healthcare (as contrasted with data collected as part of clinical trials). This resulted in the availability of the mi2b2 system. An application enabled by this technology is supported by an R01 to use clinical scans of children aged zero to six years old to build a teaching atlas.
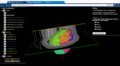
- One goal of the mi2b2 collaborative effort is to create a web-based teaching tool for fetal and neonatal anatomy as well as a browser for quantified information. A basic prototype has been created: http://fnndsc.github.io/babybrain/. The purpose of this project is to construct a 4D MRI dataset as a concatenation of multiple 3D MRI datasets (pediatric MRI scans at different stages of development), and to develop the functionality within XTK to visualize and navigate this 4D dataset in a web browser.
Approach, Plan
We propose the following steps to create a high-level didactic content creation tool.
- Enable switching between different testing scenarios (fetal brains, liver etc.)
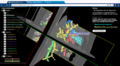
- Start with the existing Liver tutorial dataset and slides from the NA-MIC training core which includes a limited number of questions and answers
- Load a visual scene linked to a question catalog from a defined file format (XML?)
- The visualization and the questions are linked so answers can be provided in a fully interactive fashion (picking of a 3D mesh.. etc.)
- Track user-specified answers and provide statistics after testing mode
- Use 3D Slicer to generate content for training and testing
- Prototype will be built on top of existing visualization infrastructure (XTK and web-based fetal atlas tool)
- We envision a system somewhat like existing online quiz systems ([1], [2], [3], [4]) but with interactive 3D content drawn from the clinical use cases.
- May use a helper library like SlickQuiz [5]
Progress
- User interface mockups are being prepared
- Development repository: https://github.com/FNNDSC/mi2b2_aim3
- Problems/ToDo:
Update WebGLExport module to new XTK versionFix XTK with Liver CT volume (right now scalar range messes things up) or convert (normalize) the volume
- First working prototype is online!
Draft Development Plans
Phase I: Developer Mockups
During the first half of 2013:
- Hand-coded (html, javascript) mockups using to create content from existing content
- Liver tutorial from Sonia
- Fetal brain atlas
- Identify best practices and experiment with different Question and Answer modes
- Identify best practices and experiment with user interaction
- Getting feedback on mi2b2 team and refining the mockups
Phase II: Limited Beta
During second half of 2013:
- Identify first set of training content suitable for wider trial
- Set up a site to host training materials
- Set up back end system to record test results?
- Review RedCap as a back end (home page, example survey)
- Consider tie-in to RSNA Audience Response Tool
- Open site to radiology trainees and gather feedback
- Integrate infant brain atlas data as it becomes available
Phase III: Authoring Tools
During 2014:
- Develop system for clinician-educators to easily create content
- Define images, models, annotations, atlases to display
- Define didactic material
- Export content to hosting site
- Leverage tools like Scene Views and WebGL Export from Slicer