Difference between revisions of "2013 Summer Project Week:Prostate radiotherapy DICOM communication"
From NAMIC Wiki
| (One intermediate revision by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | ||
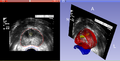
| + | Image:PW-MIT2013-Oncentra-SlicerRT.png|Oncentra test dataset in Slicer | ||
</gallery> | </gallery> | ||
| Line 8: | Line 9: | ||
* BWH: Andrey Fedorov, Emily Neubauer-Sugar, Alireza Mehrtash | * BWH: Andrey Fedorov, Emily Neubauer-Sugar, Alireza Mehrtash | ||
* Queens: Csaba Pinter | * Queens: Csaba Pinter | ||
| + | * MGH: Greg Sharp | ||
| + | * Iowa: Hans Johnson, Kent Williams | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 28: | Line 31: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Oncentra generated DICOM US can be read by Slicer, but only using nightly build (it looks like somehow nightly build is using ITK with DCMTK to read DICOM, and local builds use GDCM - this was confirmed on Mac and Windows) | |
| + | ** the issue in GDCM is that for US modality, it expects PixelSpacing to be in a different tag | ||
| + | * Oncentra generated DICOM RT can be read by SlicerRT | ||
| + | * Plastimatch standalone tools can create RT contours, but at the moment cannot reference image UIDs that correspond to 8-bit images (Plastimatch can only read images that have 16 bits allocated) | ||
| + | ** US was converted to "CT" 16-bit, corresponding RT contours created | ||
| + | * we will test the import of the generated datasets into Oncentra soon | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 13:51, 21 June 2013
Home < 2013 Summer Project Week:Prostate radiotherapy DICOM communication
Key Investigators
- BWH: Andrey Fedorov, Emily Neubauer-Sugar, Alireza Mehrtash
- Queens: Csaba Pinter
- MGH: Greg Sharp
- Iowa: Hans Johnson, Kent Williams
Objective
Our goal is to establish the capability to (1) import and display reconstructed US data and RT structures from Oncentra Prostate 4.1 system into Slicer; (2) create in Slicer multi-slice DICOM US and/or MR datasets, and DICOM RT datasets that can be imported and interpreted correctly by the Oncentra system.
Plan
- We will use the testing datasets provided with Oncentra Prostate 4.1 to test data import in Slicer and examine the specifics of DICOM structures.
- SlicerRT extensions will be used to load Oncentra RT test datasets into Slicer
- Slicer/SlicerRT/Plastimatch will be evaluated to create imaging and RT datasets, which we will attempt to import into Oncentra.
Progress
- Oncentra generated DICOM US can be read by Slicer, but only using nightly build (it looks like somehow nightly build is using ITK with DCMTK to read DICOM, and local builds use GDCM - this was confirmed on Mac and Windows)
- the issue in GDCM is that for US modality, it expects PixelSpacing to be in a different tag
- Oncentra generated DICOM RT can be read by SlicerRT
- Plastimatch standalone tools can create RT contours, but at the moment cannot reference image UIDs that correspond to 8-bit images (Plastimatch can only read images that have 16 bits allocated)
- US was converted to "CT" 16-bit, corresponding RT contours created
- we will test the import of the generated datasets into Oncentra soon