Difference between revisions of "2013 Summer Project Week:Sobolev Segmenter"
(Created page with '__NOTOC__ <gallery> Image:PW-MIT2013.png|Projects List Image:genuFAp.jpg|Scatter plot of the original FA data through the genu of the corpus…') |
|||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2013.png|[[2013_Summer_Project_Week#Projects|Projects List]] | ||
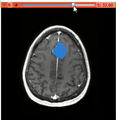
| − | Image: | + | Image:Sobolev_Segmenter2D.png|The 2D segmentation of tumor by Sobolev active contour. |
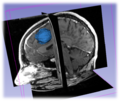
| − | Image: | + | Image:Sobolev_Segmenter3D.png|The 3D segmentation of tumor by Sobolev active contour. |
</gallery> | </gallery> | ||
| Line 16: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| + | General segmentation tools can be used in a wide range of biomedical applications from tumor delineation to segmentation of the atrial wall used in the research on the atrial fibrillation problem. | ||
| + | Sobolev active contour is a smooth general 2D segmenter that can be used in the applications above. It can be extended for medical volume segmentation. Our objective is to implement Slicer's extension based on Sobolev active contours algorithm for volume segmentation. | ||
| Line 28: | Line 30: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| + | We prepare C++ implementation of Sobolev active contour algorithm, and convert it to Slicer Commandline extension. | ||
| + | This version needs a few sparse initial contours on some slices of the segmented volume. | ||
| + | In another approach, we implement similar algorithm in Python, as an Editor effect of Slicer. In this case, only the definition of 3D depth, and a single click inside the area of the segmented target are needed for the algorithm to work. | ||
</div> | </div> | ||
| Line 35: | Line 40: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | Alpha versions of both approaches (C++ Commandline and Python Editor effect) have been prepared. | |
Revision as of 17:59, 26 May 2013
Home < 2013 Summer Project Week:Sobolev Segmenter
Key Investigators
- UAB: Arie Nakhmani, LiangJia Zhu, Allen Tannenbaum
- BWH: Yi Gao, Ron Kikinis
- Utah: Rob MacLeod, Josh Cates
Objective
General segmentation tools can be used in a wide range of biomedical applications from tumor delineation to segmentation of the atrial wall used in the research on the atrial fibrillation problem. Sobolev active contour is a smooth general 2D segmenter that can be used in the applications above. It can be extended for medical volume segmentation. Our objective is to implement Slicer's extension based on Sobolev active contours algorithm for volume segmentation.
Approach, Plan
We prepare C++ implementation of Sobolev active contour algorithm, and convert it to Slicer Commandline extension. This version needs a few sparse initial contours on some slices of the segmented volume.
In another approach, we implement similar algorithm in Python, as an Editor effect of Slicer. In this case, only the definition of 3D depth, and a single click inside the area of the segmented target are needed for the algorithm to work.
Progress
Alpha versions of both approaches (C++ Commandline and Python Editor effect) have been prepared.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a loadable Commandline extension and an Editor effect.
References
- A. Nakhmani, A. Tannenbaum, "Tracking with Adaptive Sobolev Snakes." Submitted to
IEEE Transactions on Image Processing.
- A. Nakhmani, A. Tannenbaum, "Self-Crossing Detection and Location for Parametric
Active Contours," IEEE Transactions on Image Processing, DOI:10.1109/TIP.2012.2188808, Volume 21, Issue 7, pp. 3150-3156, July 2012.