Difference between revisions of "2013 Summer Project Week:WMQL Integration in Slicer"
From NAMIC Wiki
| (2 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
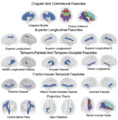
| − | Integrating the White Matter Query Language, a system for automatical definition and extraction of white matter tracts into 3D | + | Integrating the White Matter Query Language, a system for automatical definition and extraction of white matter tracts into 3D Slicer |
| Line 35: | Line 35: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | The GUI and the loadable scripted module are in place. A python console needs to be implemented in order to provide access to the interactive features of the system | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| Line 53: | Line 47: | ||
==References== | ==References== | ||
*Wassermann, et al. "On Describing White Matter Anatomy", MICCAI 2013 | *Wassermann, et al. "On Describing White Matter Anatomy", MICCAI 2013 | ||
| + | *[http://demianw.github.io/tract_querier WMQL Python implementation page] | ||
Latest revision as of 12:40, 21 June 2013
Home < 2013 Summer Project Week:WMQL Integration in SlicerKey Investigators
- BWH: Demian Wassermann, Carl-Fredrik Westin
Objective
Integrating the White Matter Query Language, a system for automatical definition and extraction of white matter tracts into 3D Slicer
Approach, Plan
We already have a pure-python toolkit to perform this. The approach would be to integrate it as a python modules
Progress
The GUI and the loadable scripted module are in place. A python console needs to be implemented in order to provide access to the interactive features of the system
Delivery Mechanism
- Slicer Module
- Extension -- Python
References
- Wassermann, et al. "On Describing White Matter Anatomy", MICCAI 2013
- WMQL Python implementation page