Difference between revisions of "2014 Project Week:TCIA Browser Extension in Slicer"
From NAMIC Wiki
| Line 30: | Line 30: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
* We developed a scripted module for Slicer which can be used to connect to TCIA archive, browse the data, download and load into Slicer scene. | * We developed a scripted module for Slicer which can be used to connect to TCIA archive, browse the data, download and load into Slicer scene. | ||
| − | * The module is finalized and is ready to be | + | * The module is finalized and is ready to be published as a Slicer extension. |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 17:08, 10 January 2014
Home < 2014 Project Week:TCIA Browser Extension in SlicerKey Investigators
- BWH: Alireza Mehrtash, Andrey Fedorov, Ron Kikinis
- Isomics: Steve Pieper
- NCI Imaging Program: Justin Kirby, John Freymann
Project Description
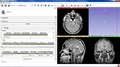
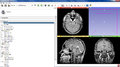
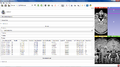
The Cancer Imaging Archive (TCIA) hosts a large collection of Cancer medical imaging data which is available to public [1]. The goal of this project is to develop a Slicer module in order to retrieve the medical images hosted at TCIA and visualize them in 3D Slicer.
Objective
- Create a scripted Slicer module for browsing TCIA collections and loading images into Slicer scene.
Approach, Plan
- Study the latest changes in TCIA API [3] and update the current module prototype.
- Discuss about the module functionality and UI design.
- Review the desired interactions of module with Slicer DICOM database.
- Review the different methods for displaying and organizing query results.
Progress
- We developed a scripted module for Slicer which can be used to connect to TCIA archive, browse the data, download and load into Slicer scene.
- The module is finalized and is ready to be published as a Slicer extension.
References
- [1] Module Code at Github: https://github.com/mehrtash/TCIABrowser/tree/master/TCIABrowser
- [2] TCIA Home Page : http://cancerimagingarchive.net/
- [3] QIICR wiki: Slicer TCIA interface project : https://github.com/QIICR/ProjectIssuesAndWiki/wiki/Slicer-tcia-interface
- [4] TCIA Rest API Documentation: https://wiki.cancerimagingarchive.net/display/Public/TCIA+Programmatic+Interface+%28REST+API%29+Usage+Guide
- [5] Module UI Design: https://docs.google.com/presentation/d/1HJcbNxhAGXP9Raw0JZFPzKXqgOgR8sv7eikYIsWPKNQ/edit?usp=sharing