Difference between revisions of "2014 Summer Project Week:TBI Segmentation"
From NAMIC Wiki
| (6 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2014.png|[[2014_Summer_Project_Week#Projects|Projects List]] | ||
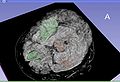
| + | Image:TBI_active_seg.jpg| Segmentation result of a subject | ||
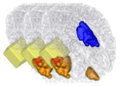
| + | Image:Active_4D.png| Illustration of user interaction | ||
</gallery> | </gallery> | ||
| Line 16: | Line 18: | ||
* The primary objective is to modify the existing code for a Slicer extension. | * The primary objective is to modify the existing code for a Slicer extension. | ||
* Determine approaches for integrating the current command line tool into Slicer 3D. | * Determine approaches for integrating the current command line tool into Slicer 3D. | ||
| + | * Understand the workflow of Python scripting module for integrating the current command line tool into Slicer 3D. | ||
</div> | </div> | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| − | <h3>Approach, Plan</h3> | + | <h3>Approach, Plan</h3> |
| − | * | + | * Try to build a basic workflow based on Slicer4Python tutorial. |
| + | * Learn from other Slicer developers for integrating the current code into Slicer. | ||
</div> | </div> | ||
| + | |||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | * | + | * Improved the instruction document of the current command line tool for interactive TBI segmentation. |
| + | * Did test for writing Python script to call C++ binaries | ||
| + | * Did test based on Slicer4Python tutorial, need to continue working on this. | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
== Reference == | == Reference == | ||
| + | * [http://www.na-mic.org/Wiki/index.php/DBP3:UCLA TBI DBP] | ||
* [http://www.nitrc.org/projects/parser_4d 4D-PARSeR (Pathological Anatomy Regression via Segmentation and Registration) Code Repo] | * [http://www.nitrc.org/projects/parser_4d 4D-PARSeR (Pathological Anatomy Regression via Segmentation and Registration) Code Repo] | ||
* [http://www.cs.utah.edu/~bowang/projects/active4D/4D_Active_Cut.html 4D Active Cut] | * [http://www.cs.utah.edu/~bowang/projects/active4D/4D_Active_Cut.html 4D Active Cut] | ||
* [http://link.springer.com/chapter/10.1007/978-3-319-02126-3_4 Modeling 4D changes in TBI] | * [http://link.springer.com/chapter/10.1007/978-3-319-02126-3_4 Modeling 4D changes in TBI] | ||
Latest revision as of 03:47, 27 June 2014
Home < 2014 Summer Project Week:TBI SegmentationKey Investigators
- GE: Marcel Prastawa
- Utah: Bo Wang, Guido Gerig
- USC: Andrei Irimia, Jack Van Horn
Project Description
Objective
- The primary objective is to modify the existing code for a Slicer extension.
- Determine approaches for integrating the current command line tool into Slicer 3D.
- Understand the workflow of Python scripting module for integrating the current command line tool into Slicer 3D.
Approach, Plan
- Try to build a basic workflow based on Slicer4Python tutorial.
- Learn from other Slicer developers for integrating the current code into Slicer.
Progress
- Improved the instruction document of the current command line tool for interactive TBI segmentation.
- Did test for writing Python script to call C++ binaries
- Did test based on Slicer4Python tutorial, need to continue working on this.