Difference between revisions of "2015 Summer Project Week:DSC"
From NAMIC Wiki
Stevedaxiao (talk | contribs) |
Stevedaxiao (talk | contribs) |
||
| Line 37: | Line 37: | ||
* [http://www.nmr.mgh.harvard.edu/~greve/dicom-unpack Dicom unpack for Freesurfer] | * [http://www.nmr.mgh.harvard.edu/~greve/dicom-unpack Dicom unpack for Freesurfer] | ||
* [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2657863/?report=reader Equations for conversion of signal to concentration for DSC] | * [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2657863/?report=reader Equations for conversion of signal to concentration for DSC] | ||
| + | * [https://github.com/stevedaxiao/DSC_Analysis.git Source code for analysis of DSC MRI] | ||
Revision as of 23:41, 23 June 2015
Home < 2015 Summer Project Week:DSCKey Investigators
- Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH), Steve Pieper (Isomics), Jayashree Kalpathy-Cramer (MGH)
Project Description
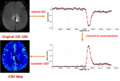
Dynamic Susceptibility Contrast (DSC) MRI imaging is an important functional imaging method that enables quantitative assessment of tissue hemodynamic patterns. Abnormality of blood flow, volume and permeability is frequently observed during tumor growth, and characterization of these perfusion attributes has become clinically important for both diagnosis and therapy planning.
Objective
- Create a module for the analysis of Dynamic Susceptibility Contrast (DSC) MRI
Approach, Plan
- Start with Slicer PKmodule
- Find the DICOM tag to identify DSC different time frame and load them as 4D data
- Update equations for conversion of signal to concentration for DSC
- Compute DSC parametric maps(eg, rCBV)
- Test some sample cases from MGH
Progress
- Equations for conversion of signal to concentration for DSC have already been updated
- Working on the parametric maps computation and DSC Dicom conversion