Difference between revisions of "2015 Summer Project Week:DSC"
From NAMIC Wiki
Stevedaxiao (talk | contribs) () |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (7 intermediate revisions by one other user not shown) | |||
| Line 21: | Line 21: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | * Start with [ | + | * Start with [https://www.slicer.org/wiki/Documentation/Nightly/Modules/PkModeling Slicer PKmodule] |
* Find the DICOM tag to identify DSC different time frame and load them as 4D data | * Find the DICOM tag to identify DSC different time frame and load them as 4D data | ||
* Update equations for conversion of signal to concentration for DSC | * Update equations for conversion of signal to concentration for DSC | ||
| Line 30: | Line 30: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
* Updated the equations for conversion of signal intensity to concentration for DSC MRI | * Updated the equations for conversion of signal intensity to concentration for DSC MRI | ||
| − | * Updated the basic equations for computing DSC parametric maps (eg, Relative | + | * Updated the basic equations for computing DSC parametric maps (eg, Relative Cerebral Blood Volume (rCBV) map) |
| − | * | + | * Discussion about loading DSC MRI Dicoms to Slicer as multi-volume 4D data |
| − | * | + | ** Read the ''Dicom to Nifti Conversion Guide of Dicom unpack for Freesurfer''[http://www.nmr.mgh.harvard.edu/~greve/dicom-unpack] |
| + | ** The tag for DSC MRI Dicoms to identify different volumes is different from DCE MRI Dicoms | ||
| + | ** Number of Volumes (ie, number of frames or time points) is the number of files with the same image position | ||
| + | ** Time between volumes/frames is the repetition time (TR) | ||
| + | ** Still debugging the code | ||
| + | * Created a module for DSC MRI analysis | ||
| + | ** Take multi-volume DSC images as input | ||
| + | ** Read Dicom header information automatically | ||
| + | ** Provide users with options to use population AIF, ROI mask, AIF mask and prescribed AIF | ||
| + | ** Output DSC parametric maps including Relative Cerebral Blood Volume (rCBV) map, Relative Cerebral Blood Flow (rCBF) map, Mean Transit Time (MTT)and etc. | ||
| + | ** Additional Options include output concentration map, fitted map and etc. | ||
| + | * Did some tests on sample data from MGH | ||
| + | * Uploaded the source code on [https://github.com/stevedaxiao/DSC_Analysis.git Github] | ||
| + | |||
</div> | </div> | ||
| Line 40: | Line 53: | ||
* [http://www.healthcare.siemens.com/siemens_hwem-hwem_ssxa_websites-context-root/wcm/idc/siemens_hwem-hwem_ssxa_websites-context-root/wcm/idc/groups/public/@global/@imaging/@mri/documents/download/mdaw/mtix/~edisp/brain_perfusion_how_why-00093544.pdf Introduction of DSC MRI from Siemens] | * [http://www.healthcare.siemens.com/siemens_hwem-hwem_ssxa_websites-context-root/wcm/idc/siemens_hwem-hwem_ssxa_websites-context-root/wcm/idc/groups/public/@global/@imaging/@mri/documents/download/mdaw/mtix/~edisp/brain_perfusion_how_why-00093544.pdf Introduction of DSC MRI from Siemens] | ||
| + | * [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2657863/ Equations for conversion of signal to concentration for DSC] | ||
| + | * [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4208985/pdf/nihms596683.pdf Other method to investigate DSC MRI] | ||
* [http://www.nmr.mgh.harvard.edu/~greve/dicom-unpack Dicom unpack for Freesurfer] | * [http://www.nmr.mgh.harvard.edu/~greve/dicom-unpack Dicom unpack for Freesurfer] | ||
| − | |||
* [https://github.com/stevedaxiao/DSC_Analysis.git Source code for analysis of DSC MRI] | * [https://github.com/stevedaxiao/DSC_Analysis.git Source code for analysis of DSC MRI] | ||
Latest revision as of 17:11, 10 July 2017
Home < 2015 Summer Project Week:DSCKey Investigators
- Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH), Steve Pieper (Isomics), Jayashree Kalpathy-Cramer (MGH)
Project Description
Dynamic Susceptibility Contrast (DSC) MRI imaging is an important functional imaging method that enables quantitative assessment of tissue hemodynamic patterns. Abnormality of blood flow, volume and permeability is frequently observed during tumor growth, and characterization of these perfusion attributes has become clinically important for both diagnosis and therapy planning.
Objective
- Create a module for the analysis of Dynamic Susceptibility Contrast (DSC) MRI
Approach, Plan
- Start with Slicer PKmodule
- Find the DICOM tag to identify DSC different time frame and load them as 4D data
- Update equations for conversion of signal to concentration for DSC
- Compute DSC parametric maps(eg, rCBV)
- Test some sample cases from MGH
Progress
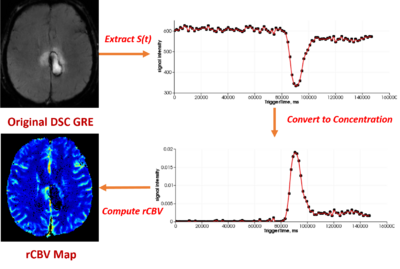
- Updated the equations for conversion of signal intensity to concentration for DSC MRI
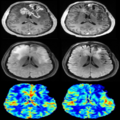
- Updated the basic equations for computing DSC parametric maps (eg, Relative Cerebral Blood Volume (rCBV) map)
- Discussion about loading DSC MRI Dicoms to Slicer as multi-volume 4D data
- Read the Dicom to Nifti Conversion Guide of Dicom unpack for Freesurfer[1]
- The tag for DSC MRI Dicoms to identify different volumes is different from DCE MRI Dicoms
- Number of Volumes (ie, number of frames or time points) is the number of files with the same image position
- Time between volumes/frames is the repetition time (TR)
- Still debugging the code
- Created a module for DSC MRI analysis
- Take multi-volume DSC images as input
- Read Dicom header information automatically
- Provide users with options to use population AIF, ROI mask, AIF mask and prescribed AIF
- Output DSC parametric maps including Relative Cerebral Blood Volume (rCBV) map, Relative Cerebral Blood Flow (rCBF) map, Mean Transit Time (MTT)and etc.
- Additional Options include output concentration map, fitted map and etc.
- Did some tests on sample data from MGH
- Uploaded the source code on Github