Difference between revisions of "2016 Summer Project Week/Brain atlas combining histology and MRI"
From NAMIC Wiki
(Created page with "==Key Investigators== * Fernando Pérez García ==Project Description== Description TODO. {| class="wikitable" ! style="text-align: left; width:27%" | Objective ! style=...") |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <gallery> | ||
| + | Image:PW-Summer2016.png|[[2016_Summer_Project_Week#Projects|Projects List]] | ||
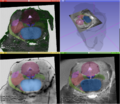
| + | Image:Brainstem contours on different images.png|Manually segmented structures of a human brainstem superimposed on a histological slice, a block-face volume and a 11.7 MRI. | ||
| + | Image:Brainstem contours 3D.png|3D visualization of the manually traced contours that have been used to interpolate the masks of the structures. | ||
| + | </gallery> | ||
| + | |||
==Key Investigators== | ==Key Investigators== | ||
| − | * Fernando Pérez García | + | * Fernando Pérez García ([http://icm-institute.org/en/ Institute of the Brain and Spine], France) |
| − | + | * Sara Fernandez Vidal ([http://icm-institute.org/en/ Institute of the Brain and Spine], France) | |
| + | * Sonia Pujol (BWH) | ||
| + | * Steve Pieper | ||
| + | * Csaba Pinter | ||
| + | * Andras Lasso | ||
| + | |||
==Project Description== | ==Project Description== | ||
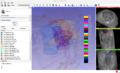
| + | Multimodal 3D visualization is important during the construction of a 3D histology / MRI atlas. Also, the interface must be very intuitive so that the user can easily trace the many contours needed for a precise 3D reconstruction. | ||
| − | + | We have chosen 3D Slicer as our development platform because of its flexibility and its huge potential for multimodal 3D visualization. Our goal is to develop an integrated, intuitive extension for manual tracing of contours on stained histological slices that have been coregistered to a high resolution MRI. | |
{| class="wikitable" | {| class="wikitable" | ||
| Line 13: | Line 25: | ||
| | | | ||
<!-- Objective bullet points --> | <!-- Objective bullet points --> | ||
| − | * | + | * A Slicer Extension to aid in the creation of a 3D MRI/histological atlas. Different modules in the extension are used for: |
| + | ** Contours tracing | ||
| + | ** Count neurons | ||
| + | ** Visualize the results | ||
| | | | ||
<!-- Approach and Plan bullet points --> | <!-- Approach and Plan bullet points --> | ||
| − | * | + | * Study the available tools in modules such as "Editor", "Segment Editor" and "Segmentations" so that they can be used in our extension. |
| + | * Create a small preview of the current histological slice (as in [https://www.researchgate.net/profile/Carlos_Ortiz-de-Solorzano/publication/233806662/figure/fig3/AS:299961800511505@1448527874676/Figure-3-Sample-histology-images-corresponding-to-a-lung-lobe-slice-of-a-mouse-17-days.png this image]), so that the user knows where they are in the slice while zooming. | ||
| | | | ||
<!-- Progress and Next steps bullet points (fill out at the end of project week) --> | <!-- Progress and Next steps bullet points (fill out at the end of project week) --> | ||
| + | * A prototype of the "Show unzoomed slice" widget has been developed ([https://youtu.be/VdOux73uWXI YouTube video]). With the help of the Slicer community, it will be improved in order to be more generic and, at some point, be integrated into Slicer's Data Probe module. | ||
| + | * A lot of feedback has been given to the developers of the "Segmentations" and "Segment Editor" modules. This will lead to improvements of these modules and will help us using their features. | ||
|} | |} | ||
Latest revision as of 15:12, 24 June 2016
Home < 2016 Summer Project Week < Brain atlas combining histology and MRIKey Investigators
- Fernando Pérez García (Institute of the Brain and Spine, France)
- Sara Fernandez Vidal (Institute of the Brain and Spine, France)
- Sonia Pujol (BWH)
- Steve Pieper
- Csaba Pinter
- Andras Lasso
Project Description
Multimodal 3D visualization is important during the construction of a 3D histology / MRI atlas. Also, the interface must be very intuitive so that the user can easily trace the many contours needed for a precise 3D reconstruction.
We have chosen 3D Slicer as our development platform because of its flexibility and its huge potential for multimodal 3D visualization. Our goal is to develop an integrated, intuitive extension for manual tracing of contours on stained histological slices that have been coregistered to a high resolution MRI.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|