Difference between revisions of "2016 Winter Project Week/Projects/BatchImageAnalysis"
From NAMIC Wiki
| Line 4: | Line 4: | ||
<!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | <!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | ||
</gallery> | </gallery> | ||
| + | |||
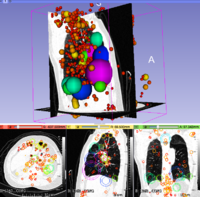
| + | [[File:LungCT-3DSIFT.png|200px|thumb|left|3D SIFT Lung Features]] | ||
==Key Investigators== | ==Key Investigators== | ||
Revision as of 19:08, 4 January 2016
Home < 2016 Winter Project Week < Projects < BatchImageAnalysisKey Investigators
- Kalli Retzepi (MGH)
- Yangming Ou (MGH)
- Matt Toews (ETS)
- Steve Pieper (BWH)
- Sandy Wells (BWH)
- Randy Gollub (MGH)
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|