Difference between revisions of "2016 Winter Project Week/Projects/ChestImagingPlatformWorkflows"
| Line 20: | Line 20: | ||
<!-- Progress bullet points --> | <!-- Progress bullet points --> | ||
* Workflows exist for particle deployment and parenchyma phenotype computation | * Workflows exist for particle deployment and parenchyma phenotype computation | ||
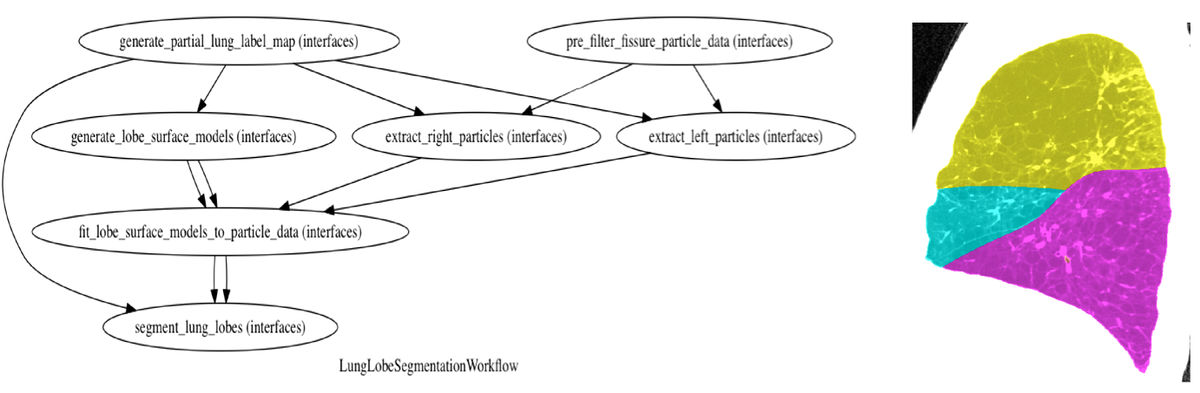
| + | * End-to-end workflow created for automatic lobe segmentation | ||
| + | * A few extra nodes to add | ||
| + | * Include in next CIP release | ||
|} | |} | ||
| − | [[File:jcrProjectWeek2016.jpg| | + | [[File:jcrProjectWeek2016.jpg|1200px]] |
Latest revision as of 02:34, 8 January 2016
Home < 2016 Winter Project Week < Projects < ChestImagingPlatformWorkflowsKey Investigators
- James Ross
- Raúl San José
Project Description
The Chest Imaging Platform (CIP) is a collection of C++ libraries, command-line executables, and python modules for segmenting, registering, processing, and quantitatively evaluating medical images of the chest; it is developed with large-scale, batch processing of high-resolution computed tomography (CT) images in mind. Many of the high-level workflows involve executing multiple CIP command-line tools and/or python modules, which imposes a barrier to entry for new users. Nipype is an open-soure, python-based software package that provides the ability to package multiple execution steps into a single workflow. In this project, I will be creating nipype-based workflows of commonly used, high-level tasks in order to make CIP functionality more accessible to new users.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|